Network pharmacology study of drug pair Tubeimu-Zhebeimu in the treatment of breast cancer
Li-Ge Gao,Tong-Jie Guo*1College of Pharmacy,Qiqihar Medical University,Qiqihar 161006,China.
#Li-Ge Gao and Tong-Jie Guo are the co-first authors of this paper.
Abstract Objective:Breast cancer is a malignant tumor endangering women’s safety and health.Clinical medication experience and related studies show that the drug pairs Tubeimu-Zhebeimu has an excellent therapeutic effect on patients with breast cancer,but its treatment mechanism is unclear.In this study,network pharmacology and molecular docking were used to analyze and explore the mechanism of“Tubeimu-Zhebeimu”in treating breast cancer.Methods:Traditional Chinese Medicine Database and Analysis Platform were used to retrieve the chemical constituents of Tubeimu and Zhebeimu,and the relevant targets were predicted through the Swiss Target Prediction Database.Searching the Gene cards,Therapeutic Target Database and Disgenet Database with the keywords“breast cancer”,“mammary cancer”and“mammary adenocarcinoma”obtain disease-related targets.We intersect the disease target with the drug target to obtain the potential drug therapy target.Then the data was imported into Cytoscape 3.9.1 software to construct a compound network of“Disease-Target-Component-Drug”,and the network.Subsequently,using the String Database a“protein-protein interaction network”was constructed and imported into Cytoscape 3.9.1 software for structural optimization and network topology analysis.DAVID was used for Gene Ontolog function enrichment and Kyoto Encyclopaedia of Genes and Genomes pathway enrichment analyses,and the results were visualized.The core targets were molecularly docked through AutoDockTools-1.5.6 software and Auto Dock Vina 1.1.2 software.Results:The results showed that the 20 active ingredients in the“Tubeimu-Zhebeimu”including β-sitosterol,Chaksine,saponins,and peimuocinine,can treat breast cancer through 139 potential targets including AKT1,AR,TP53,ESR1.Conclusion:The specific mechanism of the drug pairs Tubeimu-Zhebeimu treating breast cancer may be controlling human hormone levels,inducing cell apoptosis,and participating in the P53 protein signaling pathway and PI3K/Akt/mTOR signaling pathway.
Keywords:Tubeimu;Zhebeimu;breast cancer;network pharmacology;molecular docking;drug pairs;action mechanism
Introduction
Breast cancer is the most common cancer faced by women worldwide,and the incidence rate and mortality of breast cancer among Chinese women are also high.According to statistics,in 2019,the age group with the highest incidence rate of breast cancer is 50–69 years old,and the incidence rate is 109.19/100000,a disease that significantly endangers the safety and health of women[1].The primary treating breast cancer methods are surgery,supplemented by radiotherapy,chemotherapy,molecular targeting,and other methods[2].However,because surgical treatment depends on the onset period,and the side effects of adjuvant therapies such as chemotherapy and radiotherapy are substantial,people gradually focus on the direction of seeking drug treatment with fewer side effects and more robust efficacy.The traditional Chinese medicineTubeimuandZhebeimuhave shown a vital role in anti-tumor cancer[3].
Tubeimu,derived from the dried tubers of Bulbus Fritillaria,a cucurbitaceae plant,belongs to the Cucurbitaceae family.Its earliest written records can be traced back to the Spring and Autumn Period and the Warring States Period when it was called“Beiyi”,“meng”and so on.Zhebeimu,a plant of Liliaceae,is derived from the bulb of Fritillaria thunbergii.Its origin is Zhejiang Province.ZhebeimuandTubeimuare recorded in ancient books such as“Ben Cao Hui Yan”and“Jing Yue Quan Shu”.Zhebeimuwas initially used for anti-inflammation and analgesia[4].Later studies showed that it has pharmacological effects such as anti-oxidation,inhibiting cell proliferation,anti-acute leukemia,and reversing drug resistance of breast cancer.Xiao-Dan Zhu et al.made statistics on the prescriptions of traditional Chinese medicine for breast cancer treatment and found that the cases containing the drug pair“Tubeimu-Zhebeimu”accounted for 26.0% of the prescriptions for breast cancer and the patient’s clinical manifestations were not obvious discomfort[5].
In recent years,the experimental studies ofTubeimuandZhebeimuagainst breast cancer have increased.The research of Chao An and others showed that Tubeimoside had a strong inhibitory effect on the growth of MDA-MB-231(GFP)and MCF-7(GFP)breast cancer cells[6].In vivo experiments also showed that the extract ofTubeimuat a higher dose by oral gavage had an apparent inhibitory effect on the nude mouse model of human breast cancer MDA-MB-231-GFP[7].Li Jiansheng’s research shows thatTubeimualone can inhibit the proliferation of four different types of human breast cancer cells:MDA-MB-231,BT-549,MCF-7 and MCF-7/ADR-RES,and the addition ofZhebeimucan increase the inhibition rate of other three breast cancer cells except for MDA-MB-231 cell line[8].However,the research of“Tubeimu-Zhebeimu”against breast cancer reported in the literature only stays at the pharmacodynamics level,lacking in-depth molecular mechanism research.This paper intends to use network pharmacology to reveal the internal relationship between the drug pair and breast cancer disease and explore the molecular mechanism of the drug pair in the treatment of breast cancer through active ingredient screening,related network construction,enrichment analysis,and molecular docking technology,to provide a basis for the drug pair in the clinical treatment of breast cancer.
Materials and methods
Collection and screening of active components from Tubeimu and Zhebeimu
InputTubeimuandZhebeimuinto the Temps’Database for chemical composition query.Because Chinese herbal medicine plays a role through the practical components in the blood after oral administration,the potentially effective chemical components with high activity can be screened out according to oral bioavailability≥30% and drug-like≥0.18.
Prediction of potential targets of active ingredients
In the Pubchem Database(https://pubchem.ncbi.nlm.nih.gov),we coordinate canonical SMILES of potential practical chemical components ofTubeimuandZhebeimu,and the sorted SMILES are input into the Swiss Target Prediction Database(http://www.swisstargetprediction.ch/). The target proteins corresponding to the practical chemical components ofTubeimuandZhebeimuwere obtained,and the targets more significant than 0 were screened according to the probability index to obtain the corresponding targets of the drug to the practical chemical components.
Collection of breast cancer disease targets and acquisition of potential targets for drug treatment of breast cancer
Searching the Gene cards,Therapeutic Target Database and Disgenet Database with the keywords“breast cancer”,“mammary cancer”and“mammary adenocarcinoma”obtain disease-related targets.Map the corresponding targets of the practical chemical components ofTubeimuandZhebeimuwith the disease targets,and draw the Venn diagram.The intersection targets obtained are the potential targets for drug treatment of diseases.
Disease-Drug-Component-Intersection target network construction
The intersection targets are sorted out and then imported into the Cytoscape 3.9.1 software to construct the drug component target disease network diagram.The nodes in the network diagram represent drugs,components,diseases and target proteins respectively.The network topology is analyzed with the Network analysis plug-in in the Cytoscape 3.9.1 software.
Protein-Protein Interaction network(PPI network)construction
Import the intersection target into the STRING Database(https://string-db.org/)obtains the protein-protein interaction relationship,and then import the protein-protein interaction relationship into the Cytoscape 3.9.1 software to construct the PPI network and optimize the network to obtain the PPI network diagram of the intersection protein,analyze the network topology,and obtain the detailed topology parameters between each target.
Gene Ontology(GO)function enrichment and Kyoto Encyclopaedia of Genes and Genomes(KEGG)pathway analysis
In order to further explore the specific role of intersection targets in gene function and related signal pathways,we use the DAVID Database to perform gene ontology GO enrichment analysis on genes related to drug-disease intersection targets including four parts of molecular function,biological process,cellular component and KEGG pathway.Then it is statistically significant to set the threshold valueP<0.01,and the results are plotted into a histogram and a bubble chart for visualization by using the online drawing tool of Weisheng Xin(http://www.bioinformatics.com.cn/).
Molecular docking
To reveal the binding of the screened potential compounds to the relevant proteins in the human body after entering the human body,we performed semi-flexible molecular docking with the screened core target and the potential compounds.Download the drug 3D structure through the PubChem Database,and then modify the drug compound through AutoDockTools-1.5.6,using the modified compound as the docking ligand.Select the core target above,query and download the 3D structure in the Uniprot and PDB databases,and then import it into AutoDockTools-1.5.6 software for processing as a molecular docking receptor.Finally,molecular docking was performed with Auto Dock Vina 1.1.2 software,and the results were imported into PyMOL software for visualization.
Results
Collection and screening of active components from Tubeimu andZhebeimu
Input“Tubeimu”into the Traditional Chinese Medicine Database and Analysis Platform Database and 43 chemical components were obtained.With oral bioavailability≥30% and drug-likeness≥0.18,13 potentially active ingredients were screened out.The same method was used to queryZhebeimu,and 7 potential active components were screened out,among which β-sitosterol was a standard component ofTubeimuandZhebeimu.To facilitate the construction of the network,we numbered the potential active ingredients of the drug pair(Table 1).
Disease target screening
Searching the gene cards,Therapeutic Target Database and Disgenet Database with the keywords“breast cancer”,“mammary cancer”and“mammary adenocarcinoma”obtain disease-related targets.Then we removed duplicate targets,and 24570 breast cancer disease targets were obtained.The 24570 targets were screened according to the correlation score>10.48(double average),and 1689 disease targets were finally obtained(Table 2).
Intersection of drug target and disease target
The 19 active compounds previously predicted fromTubeimuand
Zhebeimuwere imported into the Swiss Target Prediction Database to obtain the relevant targets of each compound.The target with Probability>0 was selected,and 341 drug-related targets were obtained.The 341 drug targets were intersected with the 1689 previous breast cancer disease targets to obtain 139 intersection targets,including TP53,PIK3CA,ESR1,AKT1,EGFR,BRAF,AR,CCND1,MDM2 and other targets(Figure 1).
Disease-Drug-Component-Intersection target network
The“disease-drug-component-intersection target”network diagram ofTubeimuandZhebeimuis shown inFigure 2.This network diagram contains 447 edges and 159 nodes,of which 2 nodes represent drugs,17 nodes represent components,1 node represents diseases,and 139 nodes represent intersection targets.Topological analysis of this network shows that the main therapeutic components of the drug are ZBM-7(Degree=39),ZBM-5(Degree=37),TBM-7(Degree=29),ZBM-6(Degree=27),TBM-4(Degree=23),etc.;The main treating targets include AR(Degree=9),HMGCR(Degree=7),F2(Degree=7),SHBG(Degree=7),etc.details are shown inTable 3.

Table 3 Disease-Drug-Component-Intersection target network topology analysis(Top 10)

Figure 1 Venn diagram of the intersection of Tubeimu and Zhebeimu and breast cancer
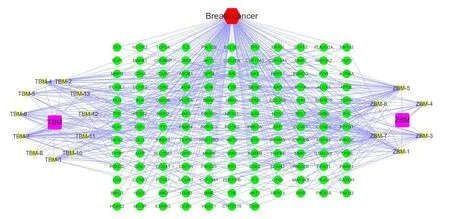
Figure 2 Disease-Drug-Component-Intersection target network.Cyan circles represent intersection targets;yellow represents components;purple squares represent drug and red hexagons represent diseases.

Table 1 19 active components from Tubeimu and Zhebeimu
Table 2 The target of breast cancer(Top 10)
PPI network
We imported 139 intersection targets into the STRING Database,and the correlation score was set to be greater than 0.7 to obtain the correlation between proteins,as shown inFigure 3(A).Import this correlation data into the Cytoscape 3.9.1 software for optimization,as shown inFigure 3(B).The network consists of 139 nodes and 2104 edges,with a moderate degree of 30.27.The size of nodes in the network is in descending order of degree value,and the color is displayed from red to green according to the degree value.In the whole PPI network,AKT1(Degree=104),TP53(Degree=102),GAPDH(Degree=94),VEGFA(Degree=90),EGFR(Degree=86),HRAS(Degree=83),SRC(Degree=81),CCND1(Degree=79)and other targets are at the core status.
Enrichment analysis
139 intersection target genes were enriched and analyzed.The results of the GO function enrichment analysis conducted through the DAVID Database showed that there were 444 GO entries in total(P<0.01),including 287 biological processes entries,75 cellular components entries,and 82 molecular functions entries,accounting for 64.6%,16.9%,18.4% respectively.According to the number of genes,the top 10 significant enrichment GO functions were selected for visualization,as shown inFigure 4.The biological process involves signal transduction,MAPK cascade,response to drugs,Fc-epsilon receptor signal pathway,etc.;cell composition involves cytosol,plasma membrane,nucleus,cytoplasm,etc.;molecular function involves protein binding,ATP binding,zinc ion binding,enzyme binding,etc.
A total of 81 signal pathways(P<0.01)were screened by KEGG pathway enrichment analysis,and the top 20 pathways ranked in the number of genes were selected for visualization(Figure 5).It involves Pathways in cancer,PI3K-Akt signaling pathway,Proteoglycans in cancer,Hepatitis B,Viral carcinogenesis,etc.In the 81 signal paths,Pathways in cancer(hsa05200)involves 54 target genes,including GSK3B,FLT3,SLC2A1,IGF1R,CCND1,etc;PI3K-Akt signaling pathway(hsa04151)involves 39 target genes,including CSF1R,GSK3B,ITGB3,PIK3CD,PIK3CB,etc;Proteoglycans in cancer involves 30 target genes,including ROCK2,SRC,ITGB3,PIK3CD,PIK3CB,etc;Hepatitis B(hsa05161)involves 29 target genes including SRC,PIK3CD,PIK3CB,TNF,PIK3CG,etc;Viral carcinogenesis(hsa05203)involves 27 target genes including HDAC2,HDAC1,SRC,PIK3CD,PIK3CB,etc.
Molecular docking
The AKT1 and TP53 in the PPI network and the AR in the“Disease-Drug-Component-Intersection target”network were selected as the receptors for molecular docking.We used the 19 potentially active components screened byTubeimuandZhebeimuas docking ligands and performed 57 molecular docking with docking receptors.The docking results are shown inTable 4,and the visualization of docking results is shown inFigure 6.Among them,β-sitosterol(TBM-1,ZBM-1)and Chaksine(ZBM-7)have excellent binding ability with AR,and the binding energy,respectively,is–9.5 kcal/mol-1and–9.7 kcal/mol-1.In order to better observe the binding of 19 components with related proteins,a molecular docking line diagram was drawn,as shown inFigure 7.

Figure 6 Docking diagram of each target and corresponding components.(A)β-sitosterol and AR;(B)Chaksine and AR;(C)TBM-12 and TP53;(D)Peimisine and TP53;(E)β-sitosterol and AKT1;(F)Ziebeimine and AKT1.

Figure 7 Line graph of molecular docking results,the abscissa is the drug component,and the ordinate is the absolute value of binding energy

Table 4 Results of molecular docking
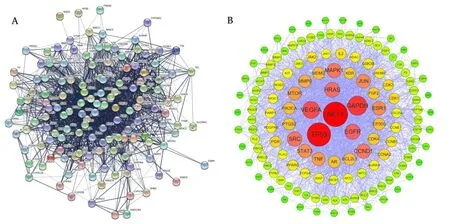
Figure 3 Protein-Protein Interaction network.(A)Protein-Protein interaction networks;(B)The colors and size of the nodes are illustrated from red to yellow in descending order of degree values.
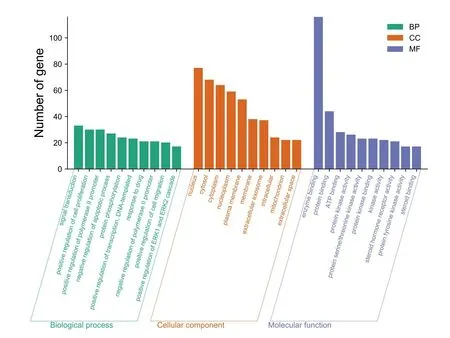
Figure 4 Analysis of GO function of Tubeimu and Zhebeimu in treating Breast cancer.GO,Gene Ontology.

Figure 5 Bubble chart of the first 20 pathways of KEGG enrichment analysis of Tubeimu and Zhebeimu target.KEGG,Kyoto Encyclopaedia of Genes and Genomes.
Discussion
The ingredients of traditional Chinese medicine are complex and diverse.The primary way to exert its efficacy is through oral administration,body circulation,participating in different biological processes,and stimulating or antagonizing specific proteins in the signal pathway,to achieve the role of treating diseases.Some studies have shown thatTubeimucan act on melanoma,lung cancer,liver cancer,colon cancer,breast cancer and other cancers,mainly by inhibiting cell proliferation,migration and invasion and inducing apoptosis to achieve the effect of cancer treatment[9].Many compounds have been isolated from Fritillaria thunbergii,including triterpenoids,sterols,alkaloids,anthraquinones,organic acids,etc.TBM-1,TBM-2,TBM-4,TBM-5 and other components belong to phytosterols and are one of the main components of animal and plant cell membranes[10,11].The human body will ingest about 300 mg of phytosterol every day,while β-sitosterol(TBM-1)has a series of pharmacological effects such as antibacterial,anti-inflammatory,anti-cancer,etc.[12–14].TBM-3,TBM-8,TBM-9,TBM-10,and other compounds belong to pentacyclic triterpenoids.Studies by Feng X and others show that Tubeimoside can induce cell cycle arrest and apoptosis[15].Zhebeimucontains polysaccharides,alkaloids,total saponins and other effective ingredients,such as Peimisine(ZBM-3)and Ziebeimine(ZBM-5),which belong to alkaloids.Wei-Yun Liu et al.showed that total nucleosides and total alkaloids ofZhebeimuinhibited the efflux activity of P-glycoprotein from drug-resistant tumor cells,and the inhibition rate was concentration-dependent[16].
This network pharmacology study screened a total of 139 potential targets of the“Tubeimu-Zhebeimu”drug pair for the treatment of breast cancer,including TP53,ESR1,AKT1,EGFR,AR,HMGCR,F2,etc.The GO enrichment analysis and KEGG pathway analysis of these 139 target proteins showed that 54 target proteins were involved in cancer regulation,39 target proteins were involved in the PI3K Akt signal pathway,30 target proteins were involved in proteoglycans in cancer,26 target proteins were involved in prostate cancer,19 target proteins were involved in estrogen signal pathway,and 18 target proteins were involved in prolactin signal pathway.It can be seen that the active ingredients of drugs in“Tubeimu-Zhebeimu”may play a role in the treatment of breast cancer by participating in the regulation of cancer signaling pathway,PI3K Akt signaling pathway,proteoglycan signaling pathway in cancer,prostate cancer signaling pathway,estrogen signaling pathway and prolactin signaling pathway.
The interaction relationship between drug components and diseases is revealed by constructing a network of“Disease-Drug-Component-Intersection Target”,in which the degree value of the AR is 9,and the proximity to centrality is 0.5,which is the core target in the network.Clinical research found that androgen has an inhibitory effect on pituitary gonadotropins,such as follicle-stimulating hormone and luteinizing hormones,thus shrinking breast cancer[17].The research results of Xiang Lu et al.show that AR can pass the Wnt/β annexin signal pathway activates HER3,which indirectly activates HER2,and effectively promotes tumor cell proliferation[18].It can also be seen from the broken line graph of molecular docking that the drug components have an excellent binding capacity with AR,and the average binding energy is–8.06kcal/mol-1.Among them,sitosterol(TBM-1,ZBM-1)and physostigmine(ZBM-7)have the most vital binding capacity with AR,which respectively are–9.5kcal/mol-1and–9.7kcal/mol-1.We speculate that there may be AR-like antagonists in the ingredients ofTubeimuandZhebeimuto achieve the therapeutic effect of breast cancer.
In the PPI network,AKT1 and TP53 are at the core status,with degrees of 104 and 102 respectively.P53 gene exists in the nucleus of each cell,but its content is shallow.Wild-type P53 is a tumor suppressor gene mainly used to induce apoptosis of cells with DNA damage.Its molecular mechanism is that damaged DNA will lead to the phosphorylation of Chk1 and Chk2 proteins[19–21].In contrast,phosphorylated Chk protein will lead to an increase in the content of P53 protein in cells,so entering and leaving the nucleus will make the Cdk2 Cyclin E complex protein inactive.The final result is that cells cannot enter the S phase from the G1 phase to apoptosis,which helps prevent the occurrence of cell carcinogenesis.The mutation of the P53 gene is often an important cause of cell carcinogenesis.The clinical research of Sun Limei and others showed that the positive rate of mutant P53 in breast cancer was as high as 44.62%,and the five-year survival rate of negative patients(65.97%)was higher than that of positive patients(52.59%)[22].Xuan-Hong Hu et al.used the immunohistochemical method to detect the expression level of Ki67,P53 antigen,and nm23 protein in 132 breast cancer tissues[23].The results showed that the expression of Ki67,P53 antigen and nm23 protein was closely related to breast cancer lymph node metastasis.The results of molecular docking and the broken line graph showed that the 19 chemical components of the drug pair had an excellent binding ability to TP53,and the average binding energy was–8.39 kcal/mol-1.Among them,the binding energy of Peimisine(ZBM-3)is–9.9 kcal/mol-1,ranking first in 57 times molecular docking.We speculate that triterpenoids,alkaloids,total saponins and other components inTubeimuandZhebeimuhave specific effects on the P53 protein,which may be mainly related to apoptosis,P53 gene mutation and other aspects.
Umemura S et al.studied the level of pAkt in different types of breast cancer patients and found that compared with breast cancer patients of other subtypes,the level of pAkt in triple-negative breast cancer patients was significantly higher,which indicated that Akt in triple-negative breast cancer was significantly activated[24].PI3K/Akt/mTOR signal pathway is an intracellular and important signal pathway[25].Research by Qiang Wu and others shows that PI3K/Akt/mTOR signal pathway can lead to tumor formation and metastasis by promoting abnormal cell differentiation,inhibiting cell apoptosis and participating in autophagy of cells[26].The PPI network topology analysis shows that AKT1 is at the network’s core with a degree value of 104 and a proximity centrality of 0.94.The results of the KEGG pathway enrichment analysis also showed that 39 of the 139 drug treatment disease targets were enriched in the PI3K-Akt signal pathway,including PIK3CD,PIK3CB,GLI1,PIK3CG,AKT2,AKT3,AKT1,AR,PIK3CA,PPARG,TP53 and other target proteins,with aPvalue of 8.03×10-33.The molecular docking of AKT1 with 19 potentially active components ofTubeimuandZhebeimushowed that AKT1 had a specific binding capacity with drug components.However,according to the broken line diagram,its binding capacity was less than TP53 and AR,and ZBM-6 had the most vital binding capacity with AKT1,which was–7.7kcal/mol-1.We speculate that the drug components may act on specific proteins in the PI3K/Akt/mTOR signaling pathway,leading to the therapeutic effect of breast cancer.
In this study,we screened the critical proteins of the drug pair“Tubeimu-Zhebeimu”in treating breast cancer through network pharmacology.We then revealed its molecular mechanism through molecular docking and a literature search.We believe that the drug treatment of“Tubeimu-Zhebeimu”mainly achieves the goal of treating breast cancer by controlling the level of human hormone,inducing apoptosis,participating in the P53 protein signaling pathway and PI3K/Akt/mTOR signaling pathway.This has provided a basis for future research on treating breast cancer byTubeimuandZhebeimu,but its specific mechanism still needs to be verified through further experiments.
 Precision Medicine Research2022年4期
Precision Medicine Research2022年4期
- Precision Medicine Research的其它文章
- Combining PD1 inhibitor,PARP inhibitor and antiangiogenic medication for lung squamous cell carcinoma with liver metastasis:a case report
- The therapeutic mechanism of Polygonatum sibiricum polysaccharide on T2DM rats based on the Nrf2 signaling pathway
- Regulatory effects of evodiamine on glucose metabolism-related factors in CT26 colorectal carcinoma-bearing mice
