Effects of Nitrogen-Regulating Gene AreA on Growth,Pathogenicity,and Fumonisin Synthesis of Fusarium proliferatum
Sun Lei, Chen Xu, #, Zhou Qianya, Zhang Tianlei, Yu Qian, Liu Lianmeng, Huang Shiwen, Wang Ling
Letter
Effects of Nitrogen-Regulating Geneon Growth,Pathogenicity,and Fumonisin Synthesis of
Sun Lei1, 2, #, Chen Xu1, #, Zhou Qianya1, Zhang Tianlei1, Yu Qian1, Liu Lianmeng2, Huang Shiwen2, Wang Ling2
(Key Laboratory of Research and Utilization of Ethnomedicinal Plant Resources of Hunan Province, College of Biological and Food Engineering, Huaihua University, Huaihua 418000, China; #)
Rice spikelet rot disease not only threatens rice yields but also poses risks to humans and animals due to the production of the category 2B carcinogen fumonisins by the pathogen. Nitrogen (N) metabolism is known to have a significant influence on fungal growth and the synthesis of secondary metabolites.is a global N regulatory gene belonging to the GATA transcription factor family. In this study, we observed that the Δmutant exhibited a notable reduction in growth rate and conidium production. Pathogenicity experiments revealed that Δhad almost lost its ability to infect rice spikelets, and become more sensitive to hydrogen peroxide (H2O2). The Δmutant could not utilize nitrate as a N source, but it could utilize ammonium (NH4+) or glutamine as a N source. The ability of Δto biosynthesize fumonisin was significantly decreased except when 120 mmol/L of ammonium chloride (NH4Cl) was used as a N source. In wild type (WT) strains, in contrast to low N sources, a high N concentration inhibited the synthesis of fumonisin B1(FB1) and the expression of fumonisin (FUM) biosynthesis genes. Conversely, Δsignificantly increased the biosynthesis of FB1at high NH4Cl concentrations and enhanced the expression of thegenes. Taken together, our results suggested thatplayed a role in positively regulating fungal growth, pathogenicity, and fumonisins biosynthesis in. NH4+can promote the FB1biosynthesis of. AreA regulates FB1biosynthesis under N starvation conditions.
The filamentous ascomycete(teleomorph) is not only the main pathogen of rice spikelet rot disease, but also simultaneously produces fumonisins, category 2B carcinogens, which have a serious impact on food safety(Huang et al, 2011a, b; Sun et al, 2019). FB1comprises almost 70% of the total amount of fumonisins produced by(Sun et al, 2019).The enzymes that catalyze the biosynthesis of fumonisins are encoded by the fumonisin biosynthetic gene cluster, which includes,,, and. All of these enzymes play key roles in the biosynthesis of fumonisin. The polyketide synthase FUM1p is a key enzyme in the first step of carbon chain assembly in fumonisins biosynthesis,encodes an aminotransferase required for the production of mature and biologically active FB1,encodes an enzyme that catalyzes the hydroxylation of fumonisin C-14 and C-15 to generate the early intermediates of fumonisins biosynthesis, and the transcription factor (TF) FUM21p can be linked to the putative TF binding motif (CGGMTA) to participate in the transcriptional activation ofand control the biosynthesis of fumonisins (Proctor et al, 2003, 2008; Shimizu et al, 2015).
Many studies have found that the metabolism of N not only affects the growth but also has an important influence on the synthesis of secondary metabolites in fungi (Tudzynski, 2014; Wei et al, 2023).Nitrogen metabolism is regulated by the GATA TF family, and preferential N sources can inhibit the expression of GATA TFs. This regulatory mechanism is called N metabolism repression (NMR). Thegene, as the main GATA transcription factor, is a core participant in the N regulatory network (Mihlan et al, 2003).has been extensively studied in many fungi. Research onhas focused on,,, and, while, to our knowledge, studies oninhave not been reported.The publicly available information ofspp. AreA/Nit-2 in the NCBI database was combined with the sequenced FP9 genome database. The sequences of genes homologous toin the FP9 strains were successfully obtained, and we successfullyconstructed deletion (Δ) and complementation (C) strains ofto investigate the effects on growth, pathogenicity, and the fumonisins biosynthesis of(Fig. 1).
Compared with WT strain FP9, the Δstrain reduced thegrowth on potato dextrose agar (PDA) media for 5 d. The amount of aerial mycelia was significantly reduced in the colony morphology with white mycelia, whereas the Cstrain significantly recovered its colony morphology after 5 d of growth(Figs. 1-A (a)–(c) and S2-A). The marginal mycelia of the Δstrain were significantly finer and had more small branches compared with the WT, while Cdid not differ significantly from the WT (Fig. 1-A (d)–(f)).The conidial yield of the Δstrain decreased significantly when grown in liquid mung bean medium (MB) for 24 h, and the conidia were significantly smaller. However, the conidial production of the Cstrain largely returned to normal (Figs. 1-A (g)–(i) and S2-B).
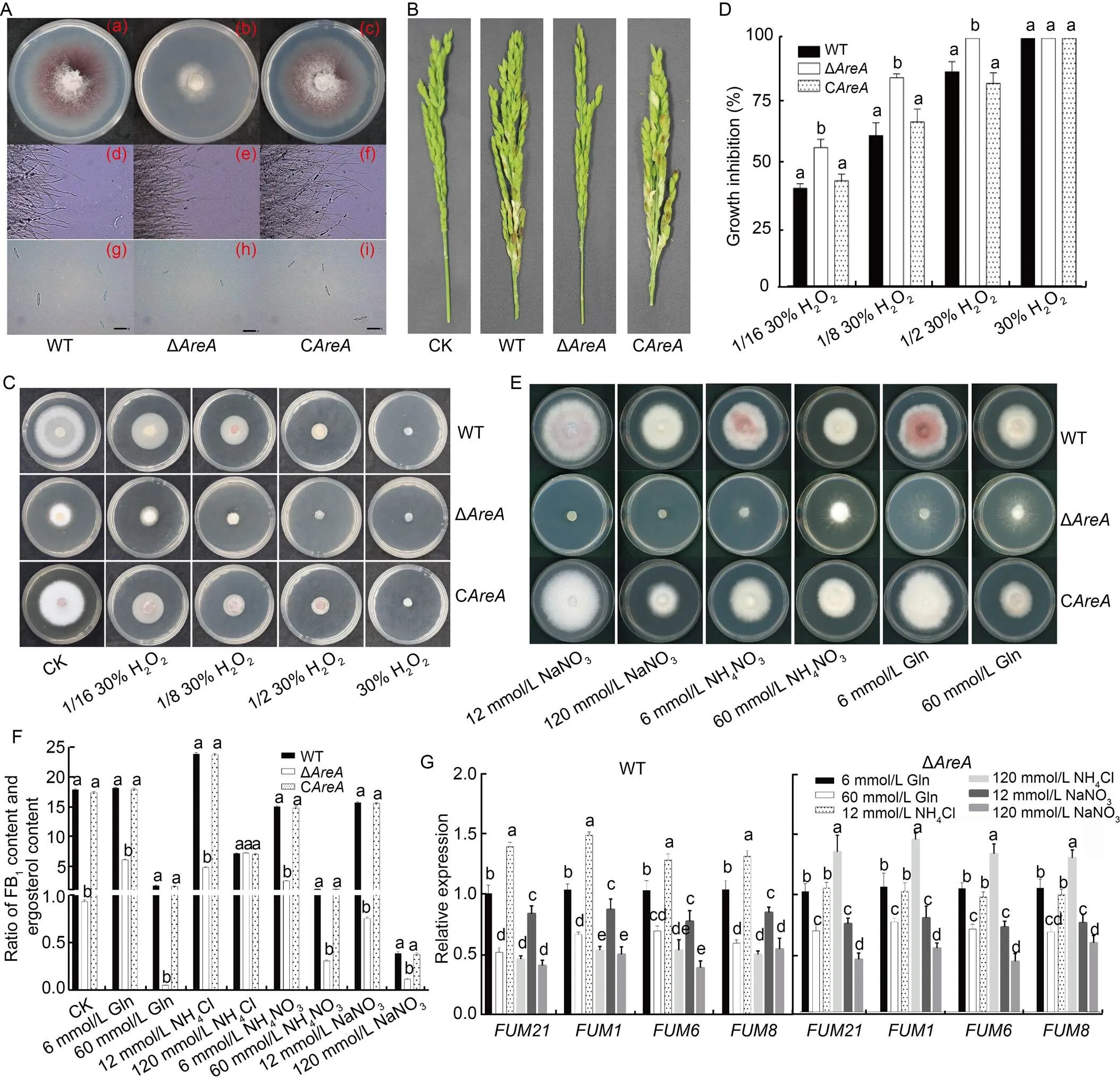
Fig. 1.Effects ofon growth, pathogenicity, and fumonisin biosynthesis of.
A, Colony morphological phenotypes of wild type (WT), deletion mutant (Δ), and complementation mutant (C) grown on potato dextrose agar (PDA) media for 5 d [(a)–(c)], morphology of mycelia at the edge of colonies of WT, Δ, and Cgrowth on PDA media for 24 h [(d)–(f)], and conidiation of WT, Δ, and Cin liquid mung bean mediafor 3 d [(g)–(i)]. Scale bars are 50 μm.B, Virulence assays were performed on rice spikelets at 14 d after inoculation. C and D, Sensitivity test (C) and growth inhibition (D) of WT, Δ, and Cstrains grown on PDA media containing different concentrations of H2O2for 3 d. E, Colony morphological phenotypes of WT, Δ, and Cgrown on minimal media with different nitrogen sources for 5 d. F, Effect of different nitrogen sources on the synthesis of fumonisins by WT, Δ, and Cstrains. G, Relative expression levels of FUM genes in WT and Δstrains in glutamine phosphate liquidmedia at 48 h with different nitrogen sources. The β-tubulin gene () was used as an endogenous control. In D, F, and G, the mean with standard deviation were calculated from three independent biological replicates. Different lowercase letters in the same graph indicate significant differences at the 5% level by the Duncan method. Gln, Glutamine.
The Δshowed severely hamperedgrowth, and the pathogenicity of the strain was inextricably linked to its growth ability. The Δand sterile water control (CK) barely caused noticeable rice spikelet rot spots on the rice glumes, while the WT and Cstrains caused noticeable symptoms (Fig. 1-B). A H2O2stress screening assay was performed to test the effect of the strain on the oxidative stress. Sensitivity of Δ, C, and WT strainsto different H2O2concentrations showed that the Δmutant was significantly more sensitive to H2O2than Cand WT, except 30% H2O2(Fig. 1-C and -D). As plants release H2O2to resist pathogen infection, the Δstrains have increased sensitivity to H2O2.Thus, the peroxide susceptibility test indicated that the Δmutant was less resistant to the host defense response, which is responsible for the reduced pathogenicity of the Δmutant.
The effect of Δstrain on the utilization of N sources was observed by growing the cultured strains on different N sources. The results showed that after 5 d of incubation in minimal media with different sources of N (NaNO3, NH4NO3, and glutamine), Δwas completely unable to grow on media with 12 mmol/L NaNO3and 120 mmol/L NaNO3. However, both the WT and Cgrew normally (Fig. 1-E). The growth rate of the Δmutant strain was significantly lower than those of the WT and Cstrain in all groups of media. Media with a high concentration of N inhibited the growth of WT and Cstrains (Fig. S3). Theinhibition was most apparent when 120 mmol/L NaNO3was used as the N source. The Δstrain grew significantly more on 60 mmol/LNH4NO3better on 6 mmol/L NH4NO3. Similarly, in the medium with glutamine (Gln) as the N source, high concentrations of the N source did not inhibit the growth of the ∆strain.
Rice grain medium, favorable for fumonisins synthesis, was used as the basic medium, and different exogenous N sources were added to investigate the ability of WT andstrains to produce FB1. The ratio of FB1amount to ergosterol content was combined to obtain the amount of FB1synthesis per unit biomass of.The ability of the Δstrain to biosynthesize FB1was significantly reduced compared with that of the WT except with 120 mmol/L NH4Cl as the N source, while the abilities of Cand WT to synthesize FB1were basically the same in all N sources (Fig. 1-F).The ability of the Cstrain to synthesize FB1was basically the same as that of the WT. Compared with the sterile water control, the addition of 12 mmol/L NH4Cl to the media used for the WT greatly increased the production of fumonisin by approximately 34.0%, while all the other groups produced less fumonisin. The FB1synthesis was severely inhibited when the WT and Cstrains were grown at a high N concentration (such as 60 mmol/L Gln, 120 mmol/L NH4Cl, 60 mmol/L NH4NO3,or 120 mmol/L NaNO3),with reductions of more than 70% in both cases. Thehigh concentration of N source inhibited the synthesis of fumonisins in the Δstrain except for the 120 mmol/L NH4Cl group.The addition of 6 mmol/L Gln, 12 mmol/L NH4Cl, 120 mmol/L NH4Cl, and 6 mmol/L NH4NO3resulted in an increase in FB1production of 552%, 416%, 674%, and 170%, respectively, in the Δstrains, compared with the sterile water control with the highest FB1content at 120 mmol/L NH4Cl.
The expression of the FUM genes in the WT was significantly higher in all media with low N concentrations compared with those with high N concentrations (Fig. 1-G). FUM expression was the highest when 12 mmol/L NH4Cl was used as the N source. In the Δstrain, except for the group with NH4Cl as the N source, the expression of FUM was also significantly higher when grown with a low N concentration rather than a high N concentration. However, when NH4Cl was used as the N source, FUM expression was higher at higher concentrations of NH4Cl compared with lower concentrations. FUM was most expressed when grown with 120 mmol/L NH4Cl. Thus, the preferred N source was more effective at activating the expression of FUM than the secondary N source at the same concentration. This result was more apparent in the ∆strain.
A comparison of the Δstrain growth on media with different N sources showed that it did not grow on media with NaNO3as the N source. In the absence of, the strain cannot utilize NaNO3, a secondary N source, and therefore, cannot grow normally. A study ofshowed that the Δstrains could not efficiently utilize secondary N sources forgrowth and have poorly developed conidiophores. Growth on complete media results in the production of significantly fewer conidia than the WT (Fasoyin et al, 2019). This is generally consistent with the results of this study. A study on the effects of different N sources on fumonisinbiosynthesis showed that a low concentration of NH4Cl promotes its biosynthesis compared with other N sources. Compared with the WT, the Δstrain produced significantly less FB1in the N source media, except in the 120 mmol/L NH4Cl group, suggesting thatpositively regulated fumonisin biosynthesis in. Similar results were obtained in studies ofin(Picot et al, 2010). Fumonisin was not synthesized in Δmutants on corn media, and the transcripts of,, andin the Δstrain were not detected by Northern analysis (Glenn et al, 2008; Kim and Woloshuk, 2008; Picot et al, 2010). Experiments with different N concentrationsshowed that high concentrations could inhibit FB1biosynthesis in both the WT and Δstrains. However, in the Δstrain, the 120 mmol/L NH4Cl did not inhibit FB1biosynthesis, instead it promoted.The results were consistent with those of studies on the effects of different N sources onFUM expression. High Nconcentrations could inhibit the expression of FUM, but the expression of FUM in the Δstrain was the highest with 120 mmol/L NH4Cl as the N source.In, the biosynthesis of deoxynivalenol (DON) by WT strains decreased significantly when 50 mmol/L NH4+was added to the rice toxin producing media, and the relative expression levels ofandwere significantly down-regulated (Hou et al, 2015). However, the relative expression levels ofandby themutant strains increased. The inhibitory effect of NH4+on DON biosynthesis should have the same effect on. The expression levels of theandgenes increased significantly inwhen grown in less than 10 mmol/L of NH4+, indicating N starvation triggered the expression of FUM genes (Kohut et al, 2009). In summary, the studies suggested thatregulates fumonisins synthesis inthrough N starvation. In the Δstrain, secondary N sources cannot be utilized, which leads to an increased demand for preferred N sources,indicating a weakened sensitivity to N concentration. Therefore, when the N source was 120 mmol/L NH4Cl, fumonisinbiosynthesis and expression levels of FUM genes were significantly increased. However, the use of a high concentration of Gln as a N source showed that it could inhibit fumonisins biosynthesis and FUM expression. The reason might be that Gln contains not only the N source but also the carbon source, and a high concentration of carbon could be the main reason for this group of experimental results. This study can provide a favorable theoretical basis not only for further analysis of the mechanism by whichaffects fumonisins biosynthesis but also reduces fumonisins contamination infrom the perspective of the utilization of different N sources.
Acknowledgements
This study was supported by the Hunan Provincial Natural Science Foundation Youth Project, China (Grant No. 2021JJ40433), and the State Key Laboratory of Rice Biology Open Project, China (Grant No. 20200301).
SUPPLEMENTAL DATA
The following materials are available in the online version of this article at http://www.sciencedirect.com/journal/rice-science; http://www.ricescience.org.
File S1. Methods.
Fig. S1. Phylogenetic tree of,reverse transplant PCR and Southern blot detection of Δand C.
Fig. S2. Growth rates of wild type, Δ, and Con potato dextrose agar media for 5 d and conidiation in liquid mung bean media for 3 d.
Fig. S3. Growth rates of wild type, Δ, and Cstrains on minimal media with different nitrogen sources for 5 d.
Fasoyin O E, Yang K L, Qiu M G, Wang B, Wang S, Wang S H. 2019. Regulation of morphology, aflatoxin production, and virulence ofby the major nitrogen regulatory gene., 11(12): 718.
Glenn A E, Zitomer N C, Zimeri A M, Williams L D, Riley R T, Proctor R H. 2008. Transformation-mediated complementation of agene cluster deletion inrestores both fumonisin production and pathogenicity on maize seedlings., 21(1): 87–97.
Hou R, Jiang C, Zheng Q, Wang C F, Xu J R. 2015. The AreA transcription factor mediates the regulation of deoxynivalenol (DON) synthesis by ammonium and cyclic adenosine monophosphate (cAMP) signalling in., 16(9): 987–999.
Huang S W, Wang L, Liu L M, Tang S Q, Zhu D F, Savary S. 2011a. Rice spikelet rot disease in China: 1. Characterization of fungi associated with the disease., 30(1): 1–9.
Huang S W, Wang L, Liu L M, Tang S Q, Zhu D F, Savary S. 2011b. Rice spikelet rot disease in China: 2. Pathogenicity tests, assessment of the importance of the disease, and preliminary evaluation of control options., 30(1): 10–17.
Kim H, Woloshuk C P. 2008. Role of, a regulator of nitrogen metabolism, during colonization of maize kernels and fumonisin biosynthesis in., 45(6): 947–953.
Kohut G, Adám AL, Fazekas B, Hornok L. 2009. N-starvation stress inducedgene expression and fumonisin production is mediated via the HOG-type MAPK pathway in., 130(1): 65–69.
Mihlan M, Homann V, Liu T W D, Tudzynski B. 2003. AREA directly mediates nitrogen regulation of gibberellin biosynthesis in, but its activity is not affected by NMR., 47(4): 975–991.
Picot A, Barreau C, Pinson-Gadais L, Caron D, Lannou C, Richard- Forget F. 2010. Factors of the-maize environment modulating fumonisin production., 36(3): 221–231.
Proctor R H, Brown D W, Plattner R D, Desjardins A E. 2003. Co- expression of 15 contiguous genes delineates a fumonisin biosynthetic gene cluster in., 38(2): 237–249.
Proctor R H, Busman M, Seo J A, Lee Y W, Plattner R D. 2008. A fumonisin biosynthetic gene cluster instrain O-1890 and the genetic basis for B versus C fumonisin production., 45(6): 1016–1026.
Shimizu K, Nakagawa H, Hashimoto R, Hagiwara D, Onji Y, Asano K, Kawamoto S, Takahashi H, Yokoyama K. 2015. The α-oxoamine synthase geneis involved in fumonisin B2 biosynthesis in., 56(3): 301–308.
Sun L, Chen X, Gao J, Zhao Y, Liu L M, Hou Y X, Wang L, Huang S W. 2019. Effects of disruption of fivegenes on fumonisin biosynthesis and pathogenicity in., 11(6): 327.
Tudzynski B. 2014. Nitrogen regulation of fungal secondary metabolism in fungi., 5: 656.
Wei Q H, Cui D Z, Zheng B J, Zhao M. 2023.antifungal activity of dihydrochelerythrine and proteomic analysis in., 30(3): 257–266.
Wang Ling (wangling03@caas.cn);
Huang Shiwen (huangshiwen@caas.cn)
13 April 2023;
6 November 2023
Copyright © 2024, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://doi.org/10.1016/j.rsci.2023.11.001
- Rice Science的其它文章
- Rice Variety Classification Based on Optimized Near-Infrared Spectral Classification Model
- Rice Husk at a Glance:From Agro-Industrial to Modern Applications
- Smart Farming for Sustainable Rice Production:An Insight into Application,Challenge, and Future Prospect
- A β-Carotene Ketolase Gene NfcrtO from Subaerial Cyanobacteria Confers Drought Tolerance in Rice
- OsbZIP01 Affects Plant Growth and Development by Regulating OsSD1 in Rice
- Potential Secretory Transporters and Biosynthetic Precursors of Biological Nitrification Inhibitor 1,9-Decanediol in Rice as Revealed by Transcriptome and Metabolome Analyses