CRISPR/Cas9系统在基因组DNA片段编辑中的应用
李金环,寿佳,吴强
上海交通大学系统生物医学研究院比较生物医学研究中心,系统生物医学教育部重点实验室,上海 200240
CRISPR/Cas9系统在基因组DNA片段编辑中的应用
李金环,寿佳,吴强
上海交通大学系统生物医学研究院比较生物医学研究中心,系统生物医学教育部重点实验室,上海 200240
源于细菌和古菌的Ⅱ型成簇规律间隔短回文重复系统[Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9),CRISPR/Cas9]近年被改造成为基因组定点编辑的新技术。由于它具有设计简单、操作方便、费用低廉等巨大优势,给遗传操作领域带来了一场革命性的改变。本文重点介绍了CRISPR/Cas9系统在基因组DNA片段靶向编辑方面的研究和应用,主要包括DNA片段的删除、反转、重复、插入和易位,这一有效的DNA片段编辑方法为研究基因功能、调控元件、组织发育和疾病发生发展提供了有力手段。本文最后展望了Ⅱ型CRISPR/Cas9系统的应用前景和其他类型CRISPR系统的应用潜力,为开展利用基因组DNA片段靶向编辑进行基因调控和功能研究提供参考。
CRISPR/Cas9系统;DNA片段编辑;基因功能;调控元件;疾病发生
随着人类基因组计划(Human Genome Project, HGP)和 DNA元件百科全书(Encyclopedia of DNA Elements, ENCODE)项目的完成,科学家们分析和鉴定了大量的人类基因组中的DNA调节元件[1~4]。这些在基因表达调控中起重要作用的 DNA调节元件包括启动子、增强子、绝缘子和沉默子等。然而很多调控元件由于遗传操作方法的限制没有得到实验的验证和功能的阐明[3, 5~10]。其次,人类基因组中包含很多串联排列、高度相似和功能冗余的基因,这些基因形成复杂的基因簇,例如原钙粘蛋白和尿苷二磷酸葡醛酸转移酶(UGT)基因簇[6, 11~13],研究基因簇的调控和功能面临巨大的挑战,急需开发遗传学基因编辑的新技术和新方法。最后,人类基因组中存在很多结构多样性(Structural variation),如DNA片段的删除、反转、重复、插入和易位[14,15],基因组结构的多样性与复杂疾病的相关性研究也多见报道[16~21]。因此有效的 DNA片段编辑方法对阐明染色体重排(Chromosomal rearrangement)和基因组结构多样性及其如何影响复杂疾病的发生发展具有重要作用。
依赖于同源重组的方法进行 DNA片段编辑的传统遗传操作,包括DNA片段的删除、反转、重复、插入和易位,均已经得到很好的发展[22~27]。近几年新出现的基因组编辑核酸酶,如锌指核酸酶(Zinc finger nucleases, ZFN)和类转录激活因子效应物核酸酶(Transcription activator-like effector nucleases, TALEN)更进一步加快了基因组编辑技术的发展[28~30]。然而这些方法在操作上技术难度大,耗时费力昂贵并且效率不高[24, 30, 31]。源于细菌和古菌的Ⅱ型成簇规律间隔短回文重复系统[Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9),CRISPR/Cas9]是新兴基因组编辑技术,由于它设计非常简单和操作方便,给遗传操作领域带来了一场革命性的改变。本文主要围绕CRISPR/Cas9系统在基因组DNA片段编辑方面的应用展开介绍。
1 CRISPR/Cas9基因组编辑系统
CRISPR/ Cas系统广泛存在于细菌和古菌中,是它们的一种适应性免疫系统,能够识别自身和外源入侵DNA片段。CRISPR/ Cas系统有3种类型:Ⅰ型、Ⅱ型和Ⅲ型[32]。Ⅰ型和Ⅲ型 CRISPR/Cas系统较为复杂,需要多个Cas蛋白形成复合体切割DNA双链,而Ⅱ型CRISPR/Cas系统只需要一个Cas9蛋白核酸酶来切割 DNA双链,即为现在广泛应用于遗传学基因编辑的CRISPR/Cas9系统。在CRISPR/Cas9系统中,Cas9核酸酶在两个非编码RNA[crRNA (CRISPR RNA)和tracrRNA (trans-activating crRNA)]的指导下直接对含有PAM (Protospacer adjacent motif)的DNA双链上游3 bp处进行靶向双链切割,形成特定位置的钝末端 DNA 双链断裂(Double strand breaks, DSBs)[33~35]。
2012年,Jinek等[36]将 CRISPR/Cas9系统中crRNA和tracrRNA两个非编码RNA改造成一个RNA,即单导向 RNA (Single-guide RNA, sgRNA),它能够指导Cas9蛋白对特定的DNA序列进行靶向断裂,为CRISPR/Cas9系统的广泛应用奠定基础。随后CRISPR/Cas9系统被应用到真核生物中,引起一场遗传操作的革命性改变。2013年,麻省理工学院张锋团队[37]和哈佛大学 Church团队[38]在Science杂志上同时发表了CRISPR/Cas9系统在哺乳动物细胞中的应用,多个靶向 sgRNAs可以同时对基因组的多个位点进行断裂,通过DNA修复系统的不精确修复在多个断裂点周围同时引发突变。但是,多个sgRNAs也可能引起意想不到的DNA片段编辑,包括可能的非常复杂的组合型(Combinatorial)DNA片段敲除、反转、重复(多个sgRNAs靶点在同一条染色体上)[39]和复杂的染色体易位(多个 sgRNAs靶点在不同的染色体上)[39]。同时 Doudna实验室[40]和Kim实验室[41]也分别报道了CRISPR在哺乳动物细胞中的应用。同年,张锋和 Jaenisch团队[42]在 Cell杂志上发表了CRISPR/Cas9系统在小鼠中的应用,进一步推动了该系统在哺乳动物中的应用研究。CRISPR/Cas9系统只需对sgRNA进行设计,具有实验简单、操作容易和节省时间等优势,已经在不同物种中迅速发展起来并得到广泛应用[43~61]。
2 CRISPR/Cas9系统在基因组DNA片段编辑中的应用
2.1 DNA片段靶向删除(Targeted deletion of DNA fragments)
研究基因和调控元件的功能,可以通过删除这一段 DNA片段来进行探索(图 1A)。2013年,CRISPR/Cas9系统被应用到哺乳动物细胞中,张锋团队[37]和 Church团队[38]发现位于同一条染色体上的两个sgRNAs对DNA双链进行操作时,在获得定点突变的同时,也存在DNA片段删除的情况(图1A)。张锋团队[37]在EMX1位点设计的相距119 bp的两个靶向sgRNAs对该119 bp DNA片段进行了删除。Church团队[38]针对AAVS1位点设计的T1和T2靶向sgRNAs,同时转染细胞后有效地获得了19 bp的DNA片段的删除。这一研究技术的出现,对阐明基因和调控元件的功能具有重要的意义。同年,Fujii 等[62]成功获得了~10 kb的DNA片段删除小鼠,并且成功繁育出下一代。同一时期,张博团队[29]成功地在斑马鱼中对DNA片段进行了删除。以上研究表明CRISPR/Cas9系统可以实现在不同物种、不同细胞系的DNA片段的删除(图1A)。
随后,不同实验室详尽报道了DNA片段删除的操作方法和效率[39, 63~65]。Canver等[63]和吴强团队[39]在哺乳动物细胞中对几十个碱基到大约一兆碱基的DNA片段进行了有效的编辑,表明此方法可以有效地对处于基因组任意位置的,任意长度的DNA片段进行操作(图 1A)。Kraft等[64]和吴强团队[39]在小鼠中对大约一千个碱基到一兆多碱基的不同长度的DNA 片段进行了有效操作,更加确定了CRISPR/Cas9系统可以在不同物种中进行任意长度的 DNA 片段操作(图 1A)。Wang等[66]通过CRISPR/Cas9系统删除了基因组中的DNA片段,鉴定了神经系统中Blimp1的增强子的存在,并阐述了增强子在Blimp1基因中的重要作用。同时在神经系统中还有其他的应用,包括对 GluN1[67]、GluA2[67]和Grin1[68]等基因的删除,表明CRISPR/Cas9系统在神经相关基因研究中的重要作用。近年来越来越多的研究表明非编码 RNA的存在可能具有重要的作用,通过 CRISPR/Cas9系统对非编码 RNA (miR-21、miR-29a和 lncRNA-21A)进行基因组操作[69],对于阐明非编码RNA的功能具有重要作用。通过删除15号染色体上大约30 Mb的DNA片段能够获得单倍体细胞系[70],为研究基因组功能和进行遗传筛选提供有用工具。这些研究表明CRISPR/Cas9系统可以实现从几十碱基对到几十兆碱基对的DNA片段的靶向删除(图1A)。
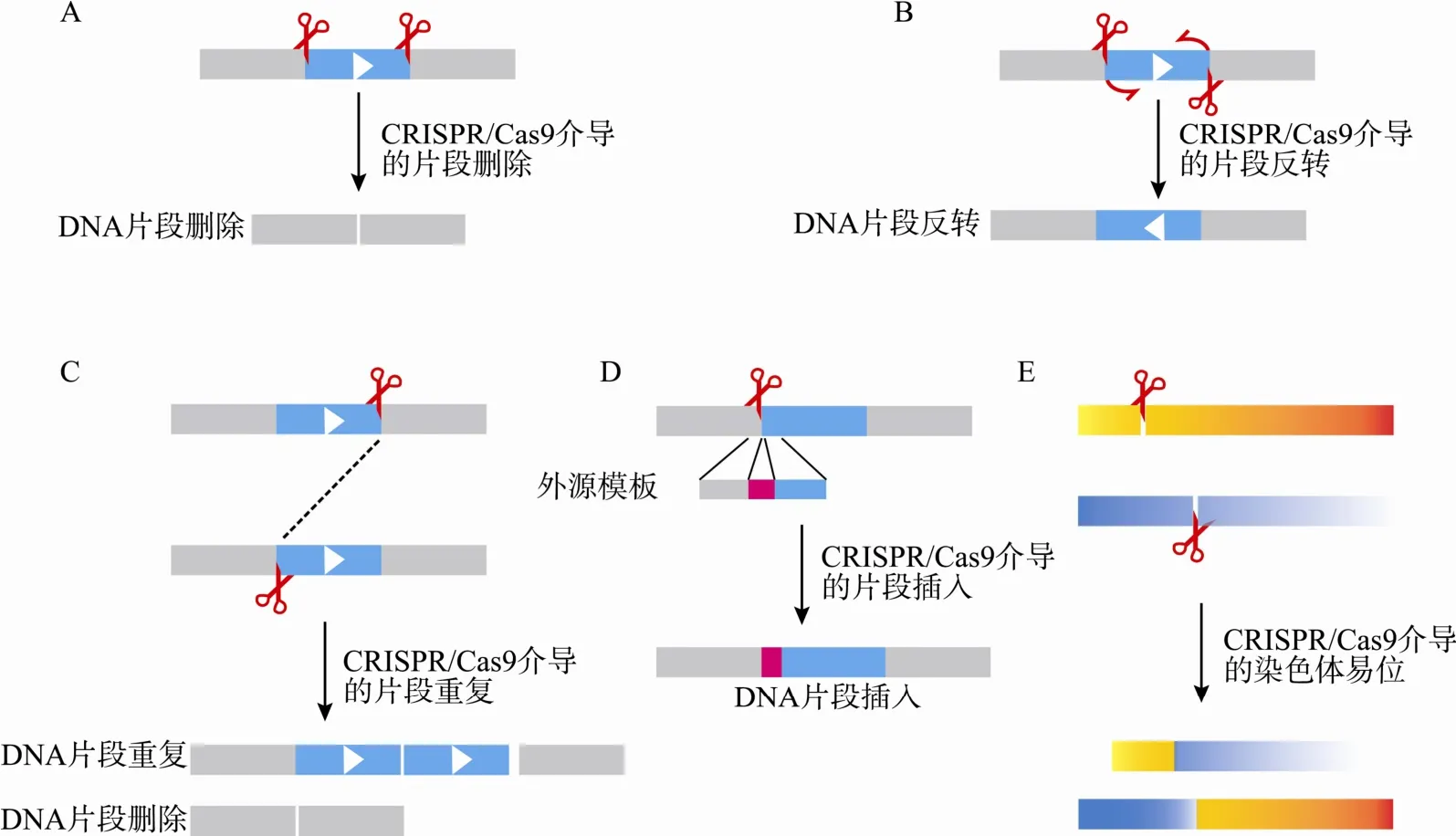
图1 CRISPR/Cas9系统在基因组DNA片段编辑中的应用
Cas9存在一定的脱靶效应[71, 72]。Ran等[47]在小鼠受精卵中通过4个靶向sgRNAs在Cas9切口酶作用下实现DNA片段的删除,他们在DYRK1A位点处成功地尝试了500~6000 bp DNA片段的删除。这种方法需要设计多一倍的靶向sgRNAs,虽然会减少脱靶率,但是多个 sgRNAs也可能引起复杂的组合型DNA片段编辑[39]。总之,利用CRISPR/Cas9系统可以实现 DNA片段的靶向删除(图 1A),尽管这一技术还有待进一步优化发展。
2.2 DNA 片段靶向反转(Targeted inversion of DNA fragments)
2014年,Choi等[73]将 CRISPR/Cas9系统应用到模拟肺癌中的基因反转事件(图1B),即EML4-ALK 和KIF5B-RET反转事件(它们导致癌症发生)。对于EML4-ALK反转事件,在人胚肾HEK293T细胞中,他们在ALK和EML4基因里分别设计一个sgRNA,大约相距12 Mb,在Cas9作用下,形成DSB,检测到了EML4-ALK反转事件,同样原理检测到大约相距11 Mb的KIF5B-RET反转事件。随后Canver等[63]在研究DNA片段删除时,也发现不同长度的DNA片段反转事件的存在,包括2~1000 kb的DNA片段反转(图1B),效率为12.8%~0.5%。同年,Maddalo 等[74]在小鼠肺组织中模拟了 EML4-ALK反转事件,他们将Cas9和两个sgRNAs一起构建到腺病毒载体上,腺病毒可以有效地感染小鼠肺,4~7周后,在感染了腺病毒的小鼠肺上会频繁地检测到肿瘤。然而这种方法不能得到可以遗传的小鼠后代,属于体细胞的改变。
2015 年,Kraft等[64]在小鼠胚胎干细胞(Embryonic stem cells, ESCs)中,通过Cas9和两个sgRNAs对1.1 kb~1.6 Mb之间的6个不同位点进行操作,检测到反转事件的存在(图 1B),然后将具有反转事件的克隆培育以获得嵌合体小鼠,该方法存在的缺陷是即使获得了反转事件的克隆,但有些克隆在后面培育小鼠的过程中并没有成功,嵌合体小鼠获得率低。吴强团队[39]在哺乳动物细胞和小鼠中对DNA片段反转进行了详尽的研究(图1B)。他们在哺乳动物细胞中对709 bp~1 Mb的7个不同长度的DNA片段进行了有效地反转(图 1B);同时他们将Cas9 mRNA和两个靶向sgRNAs注射到小鼠受精卵中,再将被注射后存活的受精卵移植到假孕鼠中获得后代,筛选得到了反转的嵌合小鼠,对960 bp~30 kb的不同长度的3个DNA片段进行了反转,并且获得了反转事件的嵌合小鼠并实现 DNA片段反转的种系传代,成功获得了F1代反转小鼠(图1B)。在人类细胞系中,他们还利用 Cas9和两个靶向sgRNAs靶向反转了7个位于不同染色体、不同大小的DNA片段(图1B)。这一简单、高效、快速的DNA片段靶向反转方法对于研究人类基因组中几万个潜在沉默子和启动子、十几万个潜在绝缘子、几十万个潜在增强子和几百万个潜在调控序列非常有用,同时也将促进对串联排列、高度相似和功能冗余的基因簇的调控和功能研究。例如,这种方法可以用来研究原钙粘蛋白基因簇中方向相反的潜在绝缘子以及其与增强子、沉默子和启动子之间的复杂关系[75]。随后吴强团队[76]利用此方法[39]反转绝缘子相关元件CBS(CTCF-binding sites)来研究基因组三维高级拓扑结构以及增强子和启动子的相互作用,阐述了CBS方向性在基因组三维高级结构和基因表达调控中的重要作用。同年,Lupiáñez等[77]根据Kraft等[64]发表的方法用小鼠建立了人类肢体发育不正常的DNA片段反转模型。通过反转DNA片段,能够改变这个 DNA反转片段所在位点周围的染色质相互作用,从而能够研究DNA结构多样性在人类肢体发育畸形中所起的重要作用。总之,利用CRISPR/Cas9系统可以实现DNA片段的靶向反转(图1B),进而研究基因表达调控和疾病发生机制。
2.3 DNA 片段靶向重复(Targeted duplication of DNA fragments)
在小鼠中能够通过基因打靶和跨等位基因重组(Trans-allelic recombination)的方法获得靶向DNA片段重复(图1C),但操作难度大,且效率偏低[27]。Kraft 等[64]在小鼠ESCs中,通过Cas9和两个靶向sgRNAs对从1.1 kb~1.6 Mb之间的6个不同位点进行操作,在其中4个位点检测到DNA重复事件的存在(图1C)然后将具有 DNA片段重复事件的克隆进行培育以获得嵌合体小鼠。吴强团队[39]在哺乳动物细胞中对709 bp~1 Mb的7个不同长度的DNA片段进行操作时,其中在5个位点检测到DNA片段靶向重复(图1C)。在小鼠中对960 bp~30 kb的不同长度的3个DNA片段进行了操作,将Cas9 mRNA和两个靶向sgRNAs注射到受精卵中,再将被注射后存活的受精卵移植到假孕鼠中获得后代,在对1241 bp DNA片段操作时,从26只删除小鼠中筛选到1只DNA片段靶向重复小鼠(图1C)。尽管利用CRISPR/cas9系统进行DNA片段靶向重复研究才刚刚起步(图1C),效率还有待提高,但这一技术将促进 CNV(Copynumber variation)的功能研究。
2.4 DNA片段靶向插入(Targeted insertion of DNA fragments)
麻省理工学院张锋团队[37]在 EMX1位点利用Cas9切口酶形成的DNA切口或Cas9形成的DNA双链断裂都可以有效地介导同源重组(homologous recombination,HR)来插入包含两个限制性酶切位点的 DNA片段(图 1D)。哈佛大学 Church团队[38]在AAVS1位点设计的T1和T2靶向sgRNAs,在Cas9和供体DNA模板作用下,通过HR可以将绿色荧光蛋白基因(Green fluorescent protein gene,GFP)内原来已经插入的终止子以及来自于AAVS1位点68 bp的片段删除,以得到正常的GFP,在此系统下他们也尝试了另外5个靶向sgRNAs,都成功的通过HR表达了绿色荧光蛋白。所以,在哺乳动物细胞中CRISPR/Cas9系统可以有效地介导HR和实现DNA片段靶向插入。同年,Wang等[42]针对 Tet1设计sgRNA,并提供能够将内源的限制性酶切位点 SacⅠ改变成为EcoRⅠ的外源DNA模板,在Cas9作用下通过 HR成功地将小鼠受精卵基因组改变,同样也针对Tet2进行操作,通过HR将内源限制性酶切位点EcoR V改变成为EcoRⅠ。更为有趣的是,他们同时对 Tet1和 Tet2进行操作,不仅获得了 Tet1和Tet2分别改变的受精卵,而且获得了Tet1和Tet2同时改变的受精卵和小鼠,表明通过 HR介导可以同时实现多位点的DNA片段靶向插入。
Byrne等[78]详尽地阐述了在人类诱导的多能干细胞中如何通过同源重组并结合 CRISPR技术进行靶向精确DNA片段插入。他们将小鼠的THY1基因靶向插入到人的 THY1基因位置并将其置换。对同源臂长度研究发现,选取同源臂长度从约100 bp~5 kb,利用CRISPR产生两个DSB时,同源臂长度在~2 kb时插入效率最高,随着同源臂长度减小同源重组效率降低,然而过长的同源臂并没有明显提高效率。对于存在一个DSB时,效率和左右两侧同源臂长度及 sgRNA靶向指导的切割位点位置有关;sgRNA切割在左侧,右侧同源臂长些时效率更高;sgRNA切割在右侧,左侧同源臂长些时效率更高。这种非对称同源臂的设计允许一侧较短的同源臂存在,可以更好地设计引物通过 PCR方法筛选克隆。Maruyama等[79]和Chu等[80]通过小分子抑制剂Scr7抑制NHEJ修复途径中的连接酶IV,大大提高了HR效率,为 HR介导的基因组编辑更广泛的应用奠定基础。总之,利用CRISPR/Cas9系统可以实现DNA片段的靶向插入(图1D)。
2.5 染色体靶向易位(Targeted translocation of Chromosomal fragments)
染色体易位常常与肿瘤发生有着密不可分的关系,因此染色体易位的研究对于人们认识肿瘤的发病机理尤为重要[81, 82]。例如,在慢性粒细胞白血病发生发展中,染色体易位造成了费城染色体(Philadelphia chromosome)形成,它能够编码BCR-ABL融合蛋白产生白血病[83]。在不同的染色体上引入同时出现的 DSBs是引发染色体易位必不可少的环节之一。利用I-Sce I或锌指核酸酶(ZFN)在染色体的特定位点引入DSBs从而引发染色体易位[84~86]。自 CRISPR技术出现以来,因其高效、便捷、易操作等特性,已有不少科研团队将其应用于染色体易位的相关研究(图1E)。
2014年,Choi等[73]利用CRISPR/Cas9技术在人胚肾HEK293T细胞和肺的上皮细胞AALE中模拟了引发肺癌的ROS1基因和CD74基因的染色体易位,说明Cas9所诱导的双链断裂能够实现染色体靶向易位。同年,Brunet团队[87]在人类细胞中利用ZFNs、TALENs和Cas9三种核酸酶,在不同的两条染色体上引入 DSBs造成染色体易位,通过对其断裂连接点的研究,发现染色体易位修复机制具有物种特异性,人类细胞和老鼠细胞利用不同的DNA修复系统。2015年,Lagutina等[88]在鼠的肌细胞中利用CRISPR系统模拟了人横纹肌肉瘤的Pax3-Foxo1基因易位事件。以上研究表明,CRISPR系统有助于人们构建与癌症相关的染色体易位模型(图 1E),从而促进癌症发病机理研究。
3 结语与展望
Ⅱ型CRISPR/Cas9系统自出现以来,得到迅速而广泛的应用,是生物学研究历史上类似PCR一样重要的技术变革。由于它具有设计简单,容易操作和成本低廉等重要优势,全世界生物学实验室都可以利用它去进行基因编辑研究。CRISPR/Cas9可以实现基因组定点编辑,在DNA修复系统的作用下实现特定位点突变[37, 38],特定位点的靶向突变可以实现基因功能的丧失。但有时在一个靶向 sgRNA和Cas9作用后所带来的突变并不能完全达到效果,这时设计两个靶向sgRNAs即可以通过DNA片段的靶向删除[29, 37~39, 62~64],有效地实现基因功能缺失。通过两个sgRNAs也可以实现DNA片段的反转和重复[39, 64, 77]。如果设计外源DNA模板,也可以通过HR实现精确的DNA靶向插入[37, 38, 78, 79]。如果靶点在不同染色体上,还可以实现DNA片段易位。所以CRISPR/Cas9系统可以实现DNA片段的删除、反转、重复、插入和易位(图1),为研究基因功能、调控元件和疾病病理机制提供有效手段。
CRISPR/Cas9系统对靶向DNA片段的编辑效率受多种因素的影响。其中,靶向 sgRNA设计对于DNA片段的编辑效率尤为重要。靶向 sgRNA序列可以选择 GC含量高一些的序列,尽量碱基分布均匀,还有靶点尽量选择在DNA超敏位点,这样可以更好地实现Cas9对DNA片段的切割[39]。DNA片段长度能够影响其删除效率[63],随着 DNA片段长度的增加 DNA片段删除效率降低[63],但片段长度也可能不影响其编辑效率[39],具体的 DNA片段编辑效率可能与基因组三维高级构象(Three dimensional higher-order architecture)相关。CRISPR系统可以实现多位点DNA编辑[37, 42],但是当多个靶向sgRNAs作用在同一条染色体上,也可能会引起非常复杂的组合型DNA片段编辑(删除、反转、重复可能会同时发生)[39],因此在用多个靶向sgRNAs进行实验时要考虑到 DNA片段编辑的复杂性。对于两个靶向sgRNAs来说,DNA片段删除和反转的效率高于DNA片段重复,DNA片段删除和反转的效率相差不大[39]。总之, 对 CRISPR/Cas9技术的研究才刚刚开始,尽管其在不同细胞类型和物种中对DNA片段的编辑效率还有待提高,但该技术一定会像其他CRISPR技术一样很快得到广泛应用。
通常CRISPR技术用来切割DNA,Ⅱ型Cas9切割 DNA必须要有 PAM位点的存在。O’Connell 等[89]设计了针对RNA切割位点的含有PAM的互补DNA寡核苷酸,发现Cas9可以切割RNA。这项研究表明CRISPR技术也可以用来研究RNA的功能。来自Francisella novicida的Ⅱ型CRISPR/Cas9也有相关报道,它可以对细菌和病毒RNA进行切割[90, 91]。Ⅰ型和Ⅱ型CRISPR/Cas系统都需要PAM位点的存在[32],即使基因组上存在很多这样的PAM位点,但
是有时在实验设计时还是会受到 PAM位点选取的限制,而Ⅲ型CRISPR/Cas系统没有PAM位点的限制,在实验设计上更加灵活。对Ⅲ型 CRISPR/Cas系统的应用目前已经有一些研究报道[92~94],在III-A 型CRISPR系统中核酸酶在crRNA(CRISPR RNA)的介导下能切割crRNA互补的DNA链和靶向DNA序列转录出来的RNA。总之,不断发展的CRISPR技术必将促进靶向DNA片段编辑应用,加速人们理解基因表达调控机制和疾病发生原因。
[1] Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC,Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H,Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature, 2001, 409(6822): 860–921.
[2] Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng ZM, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan WN, Ge WM, Gong FC, Gu ZP, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke ZX, Ketchum KA, Lai ZW, Lei YD, Li ZY, Li JY, Liang Y, Lin XY, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun JT, Wang ZY, Wang AH, Wang X, Wang J, Wei MH, Wides R, Xiao CL, Yan CH, Yao A, Ye J, Zhan M, Zhang WQ, Zhang HY, Zhao Q, Zheng LS, Zhong F, Zhong WY, Zhu SC, Zhao SY, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen MY, Wu D, Wu M, Xia A, Zandieh A, Zhu XH. The sequence of the human genome. Science, 2001, 291(5507): 1304–1351.
[3] The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature, 2012, 489(7414): 57–74.
[4] Stamatoyannopoulos JA. What does our genome encode?. Genome Res, 2012, 22(9): 1602–1611.
[5] Banerji J, Olson L, Schaffner W. A lymphocytespecific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell, 1983, 33(3): 729–740.
[6] Zhang T, Haws P, Wu Q. Multiple variable first exons: a mechanism for cell- and tissue-specific gene regulation. Genome Res, 2004, 14(1): 79–89.
[7] Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, Maurano MT, Humbert R, Rynes E, Wang H, Vong S, Lee K, Bates D, Diegel M, Roach V, Dunn D, Neri J, Schafer A, Hansen RS, Kutyavin T, Giste E, Weaver M, Canfield T, Sabo P, Zhang M, Balasundaram G, Byron R, MacCoss MJ, Akey JM, Bender MA, Groudine M, Kaul R, Stamatoyannopoulos JA. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature, 2012, 489(7414): 83–90.
[8] Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B. Amap of the cis-regulatory sequences in the mouse genome. Nature, 2012, 488(7409): 116–120.
[9] Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song LY, Vong S, Weaver M, Yan YQ, Zhang ZC, Zhang ZZ, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature, 2012, 489(7414): 75–82.
[10] de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature, 2013, 502(7472): 499–506.
[11] Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell, 1999, 97(6): 779–790.
[12] Wu Q. Comparative genomics and diversifying selection of the clustered vertebrate protocadherin genes. Genetics, 2005, 169(4): 2179–2188.
[13] Li C, Wu Q. Adaptive evolution of multiple-variable exons and structural diversity of drug-metabolizing enzymes. BMC Evol Biol, 2007, 7: 69.
[14] Sharp AJ, Cheng Z, Eichler EE. Structural variation of the human genome. Annu Rev Genomics Hum Genet, 2006, 7: 407–442.
[15] Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med, 2010, 61: 437–455.
[16] Feuk L. Inversion variants in the human genome: role in disease and genome architecture. Genome Med, 2010, 2(2): 11.
[17] Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet, 1998, 14(10): 417–422.
[18] Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet, 2005, 1(6): e49.
[19] Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature, 2007, 448(7153): 561–566.
[20] Stephens PJ, Greenman CD, Fu BY, Yang FT, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal S, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Quail MA, Burton J, Swerdlow H, Carter NP, Morsberger LA, Iacobuzio-Donahue C, Follows GA, Green AR, Flanagan AM, Stratton MR, Futreal PA, Campbell PJ. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell, 2011, 144(1): 27–40.
[21] Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, Van Allen E, Kryukov GV, Sboner A, Theurillat JP, Soong TD, Nickerson E, Auclair D, Tewari A, Beltran H, Onofrio RC, Boysen G, Guiducci C, Barbieri CE, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Ramos AH, Winckler W, Cipicchio M, Ardlie K, Kantoff PW, Berger MF, Gabriel SB, Golub TR, Meyerson M, Lander ES, Elemento O, Getz G, Demichelis F, Rubin MA, Garraway LA. Punctuated evolution of prostate cancer genomes. Cell, 2013, 153(3): 666–677.
[22] Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature, 1985, 317(6034): 230–234.
[23] Thomas KR, Capecchi MR. Introduction of homologous DNA sequences into mammalian cells induces mutations in the cognate gene. Nature, 1986, 324(6092): 34–38.
[24] Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet, 2005, 6(6): 507–512.
[25] Zheng BH, Sage M, Sheppeard EA, Jurecic V, Bradley A. Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol Cell Biol, 2000, 20(2): 648–655.
[26] Spitz F, Herkenne C, Morris MA, Duboule D. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat Genet, 2005, 37(8): 889–893.
[27] Wu S, Ying GX, Wu Q, Capecchi MR. Toward simpler and faster genome-wide mutagenesis in mice. Nat Genet, 2007, 39(7): 922–930.
[28] Gupta A, Hall VL, Kok FO, Shin M, McNulty JC, Lawson ND, Wolfe SA. Targeted chromosomal deletions and inversions in zebrafish. Genome Res, 2013, 23(6): 1008–1017.
[29] Xiao A, Wang ZX, Hu YY, Wu YD, Luo Z, Yang ZP, Zu Y, Li WY, Huang P, Tong XJ, Zhu ZY, Lin S, Zhang B.Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res, 2013, 41(14): e141.
[30] Carroll D. Genome engineering with targetable nucleases. Annu Rev Biochem, 2014, 83: 409–439.
[31] Yu YJ, Bradley A. Engineering chromosomal rearrangements in mice. Nat Rev Genet, 2001, 2(10): 780–790.
[32] Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol, 2011, 9(6): 467–477.
[33] Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology, 2009, 155(Pt 3): 733–740.
[34] Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature, 2010, 468(7320): 67–71.
[35] Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature, 2011, 471(7340): 602–607.
[36] Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 2012, 337(6096): 816–821.
[37] Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science, 2013, 339(6121): 819–823.
[38] Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science, 2013, 339(6121): 823–826.
[39] Li JH, Shou J, Guo Y, Tang YX, Wu YH, Jia ZL, Zhai YA, Chen ZF, Xu Q, Wu Q. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J Mol Cell Biol, 2015, 7(4): 284–298.
[40] Jinek M, East A, Cheng A, Lin S, Ma EB, Doudna J. RNA-programmed genome editing in human cells. eLife, 2013, 2: e00471.
[41] Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol, 2013, 31(3): 230–232.
[42] Wang HY, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell, 2013, 153(4): 910–918.
[43] Gilbert LA, Larson MH, Morsut L, Liu ZR, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell, 2013, 154(2): 442–451.
[44] Li DL, Qiu ZW, Shao YJ, Chen YT, Guan YT, Liu MZ, Li YM, Gao N, Wang LR, Lu XL, Zhao YX, Liu MY. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol, 2013, 31(8): 681–683.
[45] Malina A, Mills JR, Cencic R, Yan YF, Fraser J, Schippers LM, Paquet M, Dostie J, Pelletier J. Repurposing CRISPR/Cas9 for in situ functional assays. Genes Dev, 2013, 27(23): 2602–2614.
[46] Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 2013, 152(5): 1173–1183.
[47] Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell, 2013, 154(6): 1380–1389.
[48] Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science, 2014, 343(6166): 80–84.
[49] Wei CX, Liu JY, Yu ZS, Zhang B, Gao GJ, Jiao RJ. TALEN or Cas9-rapid, efficient and specific choices for genome modifications. J Genet Genomics, 2013, 40(6): 281–289.
[50] Cai M, Yang Y. Targeted genome editing tools for disease modeling and gene therapy. Curr Gene Ther, 2014, 14(1): 2–9.
[51] Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science, 2014, 346(6213): 1258096.
[52] González F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell, 2014, 15(2): 215–226.
[53] Harrison MM, Jenkins BV, O'Connor-Giles KM, Wildonger J. A CRISPR view of development. Genes Dev, 2014, 28(17): 1859–1872.
[54] Hsu PD, Lander ES, Zhang F. Development andapplications of CRISPR-Cas9 for genome engineering. Cell, 2014, 157(6): 1262–1278.
[55] Kiani S, Beal J, Ebrahimkhani MR, Huh J, Hall RN, Xie Z, Li YQ, Weiss R. CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat Methods, 2014, 11(7): 723–726.
[56] Niu YY, Shen B, Cui YQ, Chen YC, Wang JY, Wang L, Kang Y, Zhao XY, Si W, Li W, Xiang AP, Zhou JK, Guo XJ, Bi Y, Si CY, Hu B, Dong GY, Wang H, Zhou ZM, Li TQ, Tan T, Pu XQ, Wang F, Ji SH, Zhou Q, Huang XX, Ji WZ, Sha JH. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell, 2014, 156(4): 836–843.
[57] Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science, 2014, 343(6166): 84–87.
[58] Shen B, Zhang WS, Zhang J, Zhou JK, Wang JY, Chen L, Wang L, Hodgkins A, Iyer V, Huang XX, Skarnes WC. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods, 2014, 11(4): 399–402.
[59] Zhou YX, Zhu SY, Cai CZ, Yuan PF, Li CM, Huang YY, Wei WS. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature, 2014, 509(7501): 487–491.
[60] Wan HF, Feng CJ, Teng F, Yang SH, Hu BY, Niu YY, Xiang AP, Fang WZ, Ji WZ, Li W, Zhao XY, Zhou Q. One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res, 2015, 25(2): 258–261.
[61] Wu YX, Zhou H, Fan XY, Zhang Y, Zhang M, Wang YH, Xie ZF, Bai MZ, Yin Q, Liang D, Tang W, Liao JY, Zhou CK, Liu WJ, Zhu P, Guo HS, Pan H, Wu CL, Shi HJ, Wu LG, Tang FC, Li JS. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res, 2015, 25(1): 67–79. [62] Fujii W, Kawasaki K, Sugiura K, Naito K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res, 2013, 41(20): e187.
[63] Canver MC, Bauer DE, Dass A, Yien YY, Chung J, Masuda T, Maeda T, Paw BH, Orkin SH. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem, 2014, 289(31): 21312–21324.
[64] Kraft K, Geuer S, Will AJ, Chan WL, Paliou C, Borschiwer M, Harabula I, Wittler L, Franke M, Ibrahim DM, Kragesteen BK, Spielmann M, Mundlos S, Lupiáñez DG, Andrey G. Deletions, inversions, duplications: engineering of structural variants using CRISPR/Cas in Mice. Cell Rep, 2015, 10(5): 833–839.
[65] Li YX, Park AI, Mou HW, Colpan C, Bizhanova A, Akama-Garren E, Joshi N, Hendrickson EA, Feldser D, Yin H, Anderson DG, Jacks T, Weng ZP, Xue W. A versatile reporter system for CRISPR-mediated chromosomal rearrangements. Genome Biol, 2015, 16: 111.
[66] Wang S, Sengel C, Emerson MM, Cepko CL. A gene regulatory network controls the binary fate decision of rod and bipolar cells in the vertebrate retina. Dev Cell, 2014, 30(5): 513–527.
[67] Incontro S, Asensio CS, Edwards RH, Nicoll RA. Efficient, complete deletion of synaptic proteins using CRISPR. Neuron, 2014, 83(5): 1051–1057.
[68] Straub C, Granger AJ, Saulnier JL, Sabatini BL. CRISPR/Cas9-mediated gene knock-down in post-mitotic neurons. PLoS One, 2014, 9(8): e105584.
[69] Ho TT, Zhou NJ, Huang JG, Koirala P, Xu M, Fung R, Wu FT, Mo YY. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res, 2015, 43(3): e17.
[70] Essletzbichler P, Konopka T, Santoro F, Chen D, Gapp BV, Kralovics R, Brummelkamp TR, Nijman SMB, Bürckstummer T. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res, 2014, 24(12): 2059–2065.
[71] Fu YF, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol, 2013, 31(9): 822–826.
[72] Pattanayak V, Lin S, Guilinger JP, Ma EB, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol, 2013, 31(9): 839–843.
[73] Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun, 2014, 5: 3728.
[74] Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, Lowe SW, Ventura A. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature, 2014, 516(7531): 423–427.
[75] Guo Y, Monahan K, Wu HY, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proc Natl Acad Sci USA, 2012, 109(51):21081–21086.
[76] Guo Y, Xu Q, Canzio D, Shou J, Li JH, Gorkin DU, Jung I, Wu HY, Zhai Y, Tang YX, Lu YC, Wu YH, Jia ZL, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell, 2015, 162(4): 900–910.
[77] Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, Santos-Simarro F, Gilbert-Dussardier B, Wittler L, Borschiwer M, Haas SA, Osterwalder M, Franke M, Timmermann B, Hecht J, Spielmann M, Visel A, Mundlos S. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell, 2015, 161(5): 1012–1025.
[78] Byrne SM, Ortiz L, Mali P, Aach J, Church GM. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res, 2015, 43(3): e21.
[79] Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol, 2015, 33(5): 538–542.
[80] Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kuhn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol, 2015, 33(5): 543–548.
[81] Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer, 2007, 7(4): 233–245.
[82] Mani RS, Chinnaiyan AM. Triggers for genomic rearrangements: insights into genomic, cellular and environmental influences. Nat Rev Genet, 2010, 11(12): 819–829.
[83] Nowell PC, Hungerford DA. Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst, 1960, 25: 85–109.
[84] Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood, 2006, 107(2): 777–780.
[85] Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci USA, 2009, 106(26): 10620–10625.
[86] Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol, 2010, 17(4): 410–416.
[87] Ghezraoui H, Piganeau M, Renouf B, Renaud JB, Sallmyr A, Ruis B, Oh S, Tomkinson AE, Hendrickson EA, Giovannangeli C, Jasin M, Brunet E. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell, 2014, 55(6): 829–842.
[88] Lagutina IV, Valentine V, Picchione F, Harwood F, Valentine MB, Villarejo-Balcells B, Carvajal JJ, Grosveld GC. Modeling of the human alveolar rhabdomyosarcoma Pax3-Foxo1 chromosome translocation in mouse myoblasts using CRISPR-Cas9 nuclease. PLoS Genet, 2015, 11(2): e1004951.
[89] O'Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature, 2014, 516(7530): 263–266.
[90] Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature, 2013, 497(7448): 254–257. [91] Price AA, Sampson TR, Ratner HK, Grakoui A, Weiss DS. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc Natl Acad Sci USA, 2015, 112(19): 6164–6169.
[92] Tamulaitis G, Kazlauskiene M, Manakova E, Venclovas C, Nwokeoji AO, Dickman MJ, Horvath P, Siksnys V. Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol Cell, 2014, 56(4): 506–517.
[93] Staals RHJ, Zhu YF, Taylor DW, Kornfeld JE, Sharma K, Barendregt A, Koehorst JJ, Vlot M, Neupane N, Varossieau K, Sakamoto K, Suzuki T, Dohmae N, Yokoyama S, Schaap PJ, Urlaub H, Heck AJR, Nogales E, Doudna JA, Shinkai A, van der Oost J. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell, 2014, 56(4): 518–530.
[94] Samai P, Pyenson N, Jiang WY, Goldberg GW, Hatoum-Aslan A, Marraffini LA. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell, 2015, 161(5): 1164–1174.
(责任编委: 张博)
DNA fragment editing of genomes by CRISPR/Cas9
Jinhuan Li, Jia Shou, Qiang Wu
Key Laboratory of Systems Biomedicine (Ministry of Education), Center for Comparative Biomedicine, Institute of Systems Biomedicine, Shanghai Jiao Tong University, Shanghai 200240, China
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9) system from bacteria and archaea emerged recently as a new powerful technology of genome editing in virtually any organism. Due to its simplicity and cost effectiveness, a revolutionary change of genetics has occurred. Here, we summarize the recent development of DNA fragment editing methods by CRISPR/Cas9 and describe targeted DNA fragment deletions, inversions, duplications, insertions, and translocations. The efficient method of DNA fragment editing provides a powerful tool for studying gene function, regulatory elements, tissue development, and disease progression. Finally, we discuss the prospects of CRISPR/Cas9 system and the potential applications of other types of CRISPR system.
CRISPR/Cas9 system; DNA fragment editing; gene function; regulatory elements; diseases
2015-06-23;
2015-08-27
国家自然科学基金项目(编号:31171015,31470820)和上海市科学技术委员会(编号:13XD1402000,14JC1403600)资助
李金环,博士研究生,专业方向:遗传学。 E-mail: lijinhuan163@126.com 寿 佳,博士研究生,专业方向:遗传学。 E-mail: shoujia1106@163.com李金环和寿佳同为第一作者
吴强,博士,教授,博士生导师,研究方向:基因表达调控及神经发育。E-mail: qwu123@gmail.com
10.16288/j.yczz.15-291
时间:2015-9-1 9:32:39
URL:http://www.cnki.net/kcms/detail/11.1913.R.20150901.0932.002.html

