Decoding epigenetic codes: new frontiers in exploring recovery from spinal cord injury
Bo-Yin Zhang, Peng-Yu Chang , Qing-San Zhu, Yu-Hang Zhu, Saijilafu
1 Department of Orthopedic Surgery, China-Japan Union Hospital of Jilin University, Changchun, Jilin Province, China
2 Department of Radiotherapy, The First Bethune Hospital of Jilin University, Changchun, Jilin Province, China
3 Department of Orthopedics, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu Province, China
Abstract Spinal cord injury that results in severe neurological disability is often incurable. The poor clinical outcome of spinal cord injury is mainly caused by the failure to reconstruct the injured neural circuits. Several intrinsic and extrinsic determinants contribute to this inability to reconnect. Epigenetic regulation acts as the driving force for multiple pathological and physiological processes in the central nervous system by modulating the expression of certain critical genes. Recent studies have demonstrated that post-SCI alteration of epigenetic landmarks is strongly associated with axon regeneration, glial activation and neurogenesis. These findings not only establish a theoretical foundation for further exploration of spinal cord injury, but also provide new avenues for the clinical treatment of spinal cord injury. This review focuses on the epigenetic regulation in axon regeneration and secondary spinal cord injury. Together, these discoveries are a selection of epigenetic-based prognosis biomarkers and attractive therapeutic targets in the treatment of spinal cord injury.
Key Words: axon regeneration; epigenetic biomarkers; epigenetic modification; histone; microRNA; prognosis; secondary injury; spinal cord injury
Introduction
When spinal cord injury (SCI) results in a catastrophic attack on the central nervous system (CNS), it can lead to permanent limb paralysis or even cardiorespiratory failure. With the expansion in the transportation and construction industries, the incidence and prevalence of SCI have increased steadily. The estimated incidence rate in the United States was 54 cases per million in 2012 (Jain et al., 2015). The rate was slightly lower in China at 37 cases per million. Even so, around 20.5% cases were classified as American Spinal Injury Association grade A and the incomplete paraplegia rate was 27.9% in 2016 (Chen et al., 2018; Li et al., 2019). Consequently, the high incidence and morbidity rates of SCI have become a significant challenge for society and the medical system. Unfortunately, SCI induced neurological disability is currently incurable (Courtine and Sofroniew, 2019).
SCI is a clinical syndrome that consists of multiple pathological processes, and can be divided chronologically into three continuous phases: acute, sub-acute and chronic phases (Ahuja et al., 2017). Pathophysiologically, the devastating neural function consequence of SCI can be attributed to both the primary and secondary injuries of SCI (McDonald and Sadowsky, 2002; Ramer et al., 2014). The primary injury is the mechanical attack on the spinal cord, which directly destroys the integrity of neural circuits of the spinal cord. In this stage, neuroglial cells start to proliferate and limit the lesion from spreading (Barnabé-Heider and Frisén, 2008). Afterwards, stress signaling crosstalk activation and inf lammatory cascades trigger the onset of the secondary injury. Secondary injury-induced growth inhibitory molecules, such as chondroitin sulfate proteoglycans and myelin-associated proteins, impede axon re-projection across the glial-rich lesion (Orr and Gensel, 2017). In addition, the post-injury massive inflammatory reaction aggravates the neuron and oligodendrocyte loss and eventually represses neuronal survival in the lesion. Pathologically, internal and external barriers are responsible for the poor functional recovery post-SCI (Vogelaar, 2016). Intrinsically, the poor regenerative responses of cleaved axons hinder the reconstruction of neural connections. Post SCI, both the demyelination and stump degeneration of axons destroy the neuronal network. In addition, compared with precursor cells, the regeneration potential of mature neurons is limited. Extrinsically, glial activation and inf lammatory inf iltration turn the spinal lesion site into a hostile environment for the regeneration of neurons (Hur et al., 2012). Therefore, restoration of neural function post-SCI requires the favorable coordination of intrinsic neuronal regenerative capacity and the unlocking and coordination of various extrinsic elements (Figure 1).
Epigenetics refers to the regulation of gene expression, orchestrated by chromatin modifications, DNA structure alteration and non-coding of RNAs independent of the DNA sequence transformation (Christopher et al., 2017). In the CNS, emerging evidence demonstrates that epigenetic regulation plays a critical part in multiple pathological and physiological processes, such as proliferation, differentiation, survival and regeneration (Kameda et al., 2018; Sfera et al., 2018). SCI is accompanied by a series of epigenetic landmark alterations, some strongly related to processes such as axon regeneration, glial activation, neurogenesis and ependymal cell reprogramming (Li et al., 2016). By recognizing these epigenetic changes, the SCI associated extracellular stress signal can be translated into intracellular signals readable by endogenous spinal cord cells. Consequently, decoding the regulatory controls behind these post-SCI epigenetic changes is of great importance to enable development of clinical solutions that promote the mechanisms of SCI recovery (Figure 2 and Table 1). Fortunately, technological improvements in epigenetics-associated sequencing and drug delivery have brought these experimental breakthroughs closer to clinical SCI therapy (Zhang et al., 2013; Li and Rana, 2014).
This review summarizes the past two decades of major progress in the understanding of epigenetic regulation in axon regeneration during the secondary recovery phase after SCI. It also focuses on the selection of epigenetics-based prognostic biomarkers and the clinical trials of SCI treatment. The articles published prior to November 2019 were retrieved in PubMed database with the following terms: spinal cord injury; epigenetic modification; axon regeneration; secondary injury; prognosis; histone; microRNA; epigenetic biomarkers.
Epigenetic Modifications and Axon Regeneration Awakening
As a structural and functional unit of a neural circuit, the axon plays a central role in nerve signal transmission. Therefore, injuries in the CNS or the peripheral nervous system require axon regeneration for functional neurological recovery. Unfortunately, in the mammalian CNS, particularly in the spinal cord, most axons exhibit extremely limited regenerative capacity post-injury (Li et al., 2016). The underlying mechanisms, however, remain unknown. Post-injury soma gene expression, such as recruitment of regeneration associated genes (RAGs) and the output of cytoskeleton components, are major determinants of axonal growth capacity (Hur et al., 2012). Previous studies have proposed that poor RAG activation in response to injury is the reason for the failure of axon regeneration post-SCI (Li et al., 2016). By comparison, during development, the axons and dendrites of newborn neurons possess the growth capacity to support those processes to project to their destinations. Once mature, the ability of axons to grow declines and the neurons mainly form new synapses and produce neurotransmitters (Kameda et al., 2018). Physiologically, the developmental process from primitive stage to terminal state of neuron is irreversible. Therefore, the loss of regenerative capacity can be regarded as the cost of acquiring functionality. Fortunately, as an exception to this predicament, the peripheral axotomy provides a silver lining for SCI study. Axotomy triggers the expression of several RAGs in the soma, inducing a vigorous regenerative response that substantially promotes the regeneration of the damaged axon, almost as if the neurons had regressed to the development phase (Palmisano and Di Giovanni, 2018). Consequently, improving the expression of RAG in impaired neurons is the key to unlock this regeneration capability (Palmisano et al., 2019). Recently, studies have demonstrated that several epigenetic regulations manipulate the injured axon reprogramming by modulating RAG expression.
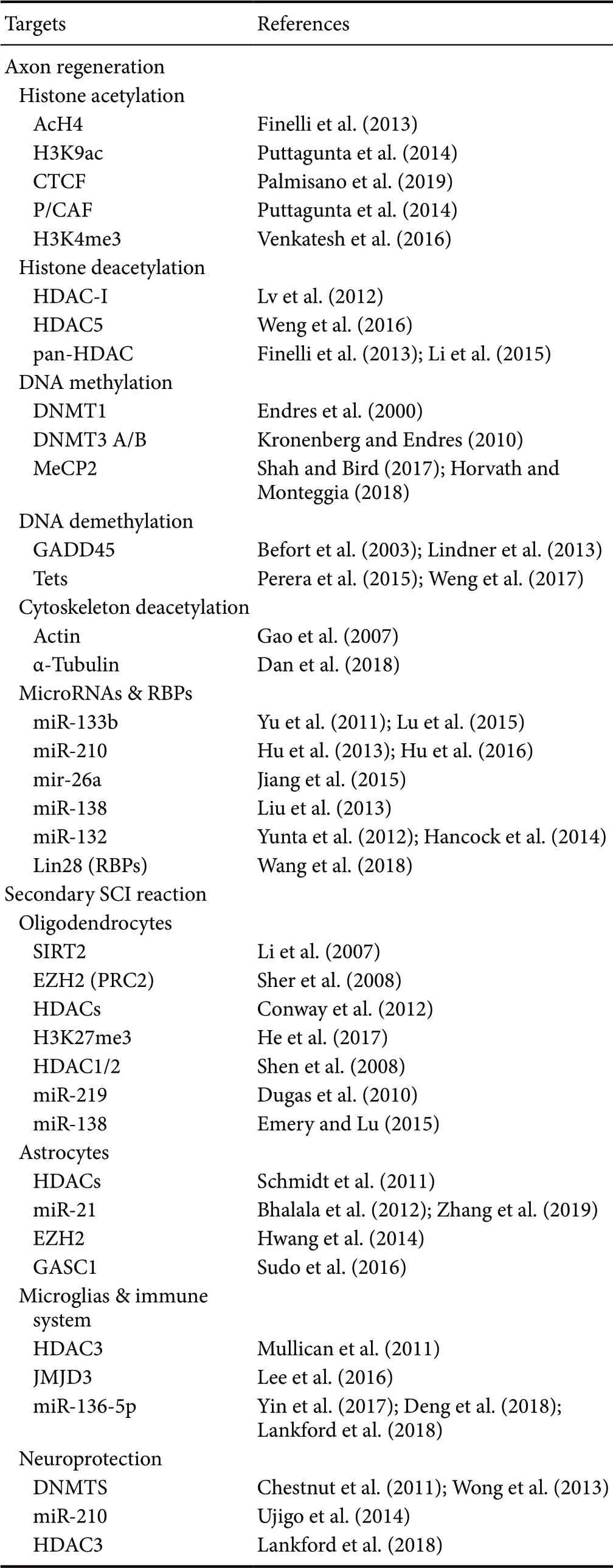
Table 1 Potential neuroepigenetic targets of spinal cord injury

Figure 1 Pathological changes and cellular responses post-spinal cord injury (SCI).

Figure 2 Deciphering spinal cord injury associated epigenetic codes.
Histone modifications and axon regeneration
The three-dimensional conformation of chromatin directly determines DNA accessibility and therefore its gene expression status. The histone modifications, also known as histone codes, on amino-terminal tail lysine residues are flexibly edited by covalent transferases that are critical regulators of chromatin remodeling (Maury and Hashizume, 2017). Recent discoveries have demonstrated the close relationship between histone codes and RAG expression post nerve injury.
Histone acetylation associated modifications
Histone acetylation is under the coupling regulation of histone acetyltransferases (HAT) and histone deacetylases (HDACs). HATs increase histone hydrophobicity by transferring the active acetyl group from coenzyme A to lysine residues in the histone. The acetylated chromatin converts to a less compact state that promotes gene expression. In contrast, HDACs exerts a gene silencing effect during transcription (Stam et al., 2007). For matured neurons, a lower histone acetylation level is beneficial to maintain structural and functional stability. However, this stability usually reduces the neuron repair capacity.
Recent studies have suggested that the poor regenerative performance of axons is probably caused by the H4 hypoacetylation state that inhibits the axon reprogramming post-injury. However, a peripheral axotomy triggers an up-regulation of H4 acetylation, which leads to AcH4 enrichment at RAGs promoter regions (Finelli et al., 2013). H3K9 acetylation modifications also play a pivotal role in the regulation of RAGs expression. A sciatic nerve axotomy increases the active epigenetic marker H3K9ac and decreases the repressive epigenetic marker H3K9me2. The H3K9ac is then recruited at the promoter sites of RAGs, such as gap43, galanin, and bdnf (Puttagunta et al., 2014). Furthermore, the sequence-specific DNA-binding protein CTCF (CCCTC-binding factor) is recognized as an essential neuroepigenetic element of peripheral conditional lesion-induced axon regeneration (Palmisano et al., 2019). SCI also promotes sensory axon regeneration by facilitating RAG expression through exogenous overexpression of acetyltransferases, such as p300/CBP-associated factor (P/CAF) (Puttagunta et al., 2014).
In a rodent SCI model the locomotor function could be profoundly accelerated by employing the HDAC I inhibitor valproic acid post-injury. Mechanically, the valproic acid-induced a hyperacetylation condition that not only promotes glial cell line-derived neurotrophic factor and brain-derived neurotrophic factor (BDNF) expression, but also weakens the axon tip recognition of Nogo-A inhibitory signals (Lv et al., 2012). Similarly, the artif icial elevation of histone acetylation levels by applying HDACs inhibitors, such as trichostatin A and MS-275, can largely accelerate RAGs expression and sensory axon regeneration post-injury (Finelli et al., 2013). As well as histone residues, axon cytoskeleton components also serve as substrates of HDACs (Li et al., 2015). The traumatically induced calcium inf lux translocates phosphorylated PKCu from the cytoplasm into the nucleus, which in turn exports HDAC5 to the cytoplasm. The exported HDAC5 was portaged to the growth cone via axonal transportation. By deacetylating the microtubules in a growth cone, HDAC5 enhances the cytoskeletal dynamics, and promotes axon regeneration (Weng et al., 2016). In addition, Gao et al. (2007) demonstrated that HDAC6 orchestrates the cytoskeletal remodeling in a actin-based manner. As well as actin, another axon cytoskeleton component, α-tubulin, acts as a substrate for acetylation modification. Specif ically, α-tubulin acetylation turns microtubules into a hypo-dynamic state, this then acts as a molecular brake on axon regrowth and over branching during development (Dan et al., 2018).
Histone methylation associated modifications
Histone methylation is another typical epigenetic modification that regulates chromatin accessibility. One of the most remarkable characteristics of histone methylation modification is the regulation of bivalent domains (Swigut and Wysocka, 2007). Structurally, the bivalent domain is shaped by two components, the negative unit H3K27me3, and the positive unit H3K4me3. These units exert conjugated regulation on the expression of certain genes. Consequently, any factors that break the bivalent domain balance will inf luence the modif ied gene expression status. A recent study in E15 cortical neurons suggests that H3K4me3 is enriched at the promoter regions of RAGs, such as gap43, sprr1a, integrina7, and galanin, that play a positive role in gene expression. However, the H3K4me3 enrichment at RAGs promoter sites was replaced by H3K27me3 after maturity (Venkatesh et al., 2016). Accordingly, the poor growth capacity of mature neurons is in part due to the inhibition of RAGs gene expression by H3K27me3. Although direct evidence of axon regeneration related histone methylation modification is yet to be explored, it might be worthwhile system to investigate as a possible check on the effectiveness of future treatment for SCI.
DNA modifications and axon regeneration
To maintain the stability of the mature nerve system, differentiated neurons are bound by inhibitory epigenetic marks, which turn the neuron into a conservative state. Although those marks can guarantee the stable performance of neurons, they deprive the mature neurons of the capability for robust growth. DNA methylation is one of the key epigenetic codes that inhibit axon regeneration. Under the catalysis by DNA methyltransferase family enzymes (DNMTs), the 5-methylcytosine is deposited at cytosine-phosphate-guanine (CpG) dinucleotides when an active methyl group is added to cytosine nucleotides of a DNA single strand. Notably, DNA methylation is under reversible regulation by DNMTs and demethylation proteins, which can add and remove methylation groups during transcription (Zou et al., 2009). Accordingly, deciphering and manipulating those epigenetic codes in mature neurons will raise the possibility of crossing the intrinsic barrier to axon regeneration.
DNA methylation
Folate-mediated DNA modification was early evidence of axon-associated DNA methylation. The folate, also named vitamin B9, was originally discovered as a donor of single-carbon metabolism (Balaghi and Wagner, 1993). In the sciatic transection model, there is a sharp up-regulation of the folate receptor-1 in lesions, which enhances the folate uptake capacity of injured neurons (Iskandar et al., 2010). Both DNMT3A and B expression and global DNA methylation are simultaneously and dramatically decreased post nerve injury. Based on this evidence, the nerve-related therapeutic effects of folate mediated DNA modification were further explored. By employing exogenous folate, the DNMT3A and B protein expression of injured nerves was restored, and the de novo DNA methylation was maintained. Importantly, the axon regeneration was largely promoted under the abundant folate state, which supplied convincing evidence for its clinical application (Kronenberg and Endres, 2010). Post ischemic CNS injury, the DNMT1-dependent DNA methylation increased in the lesion. Selective knockout of DNMT1, other than pan DNMT, def initely contributes to the synapse formation and neuron survival (Endres et al., 2000). It is paradoxical that subunits of DNMTs family, such as DNMT1 and DNMT3A/B, exhibit distinct responses to nerve injury stress. Functionally, DNMT3 regulates de novo DNA methylation, while DNMT1 is responsible for maintaining DNA methylation levels, especially during DNA replication. The diversity of gene capture among DNMTs subunits might be a possible explanation for their functional difference. For instance, the inactivation of DNMT1 is likely to unlock the RAGs, which were restrained by the stable DNA methylation. More investigations are still necessary to determine the DNMTs’ regulation network. In addition to regulation of axonal regeneration, DNMTs were demonstrated to mediate motor neuron death in neurological diseases such as amyotrophic lateral sclerosis (Wong et al., 2013). In patients with amyotrophic lateral sclerosis, the DNMT up-regulation induced aberrant DNA methylation in motor neurons; this may indicate a possible mechanism of motor neuron apoptosis and neurodegeneration (Chestnut et al., 2011). Consequently, more investigations are still necessary to determine the DNMTs’ regulation mechanisms in nerve systems to balance the benef its and detriments of DNMTs post-SCI.
Methyl CpG-binding protein 2 (MeCP2) is a methyl-CpG-binding protein that acts as a reader for DNA methylation. MeCP2 expression is enriched in CNS neurons and the mutation of MeCP2 is the molecular etiology for the X-linked dominant disorder Rett syndrome (Shah and Bird, 2017). Earlier studies have recognized MeCP2 as a pure transcription repressor (Martinowich et al., 2003). One of the well-known direct downstream targets of MeCP2 is BDNF; which is critical for neuron survival, synaptogenesis, and axon outgrowth (Chen et al., 2003). The binding of MeCP2 and Bdnf promoter leads to suppression of BDNF expression. Neuronal stimulation-induced MeCP2 phosphorylation can expose the Bdnf promoter and facilitate BDNF synthesis. However, besides its repressor property, MeCP2 has been shown to act as an activator of gene expression (Horvath and Monteggia, 2018). Chahrour et al. (2008) examined the gene expression prof iles in MeCP2 overexpression conditions and found that thousands of gene expressions were up-regulated post MeCP2 overexpression. In addition, MeCP2 was detected to form a positive transcription complex with cAMP response element-binding 1, a positive regulator of RAGs, in the brain during transcription (Lindner et al., 2013). These findings not only deepen the understanding of the molecular mechanisms of Rett syndrome-associated neurological disorders but provide new insights into axon regeneration.
DNA demethylation
DNA methylation markers can be removed by certain DNA demethylation proteins. Ten-eleven translocation (Tet) methylcytosine dioxygenase can oxidize the 5-methylcytosine to 5-hydroxymethylcytosine (5-hmC) in DNA, which acts as a DNA demethylation protein during transcription. In the postnatal period, the constant accumulation of 5hmC induced by Tet mediated oxidation was seen to be correlated to retinal cell differentiation and axon outgrowth (Perera et al., 2015). Weng et al. (2017) discovered that the lesioning of the peripheral nerve induced axon regeneration that was parallel to the up-regulation of Tet3 expression in dorsal root ganglion neurons. Certain RAGs, such as Atf3, which were previously blocked by DNA methylation, were unlocked by post-injury induced Tet3 DNA demethylation (Weng et al., 2017). GADD45 has DNA demethylation activity and is important for post damage DNA repair (Barreto et al., 2007). In both the sensory and motor neurons, GADD45 expression is sharply increased post nerve injury (Befort et al., 2003). In addition, the intracellular distribution of GADD45 is consistent with RAGs such as c-Jun (Lindner et al., 2013). Based on this evidence, the potential relationships between DNA demethylation proteins and axon regeneration are worth further exploration.
MicroRNAs and axon regeneration
MicroRNAs (miRs) are a class of small non-coding RNAs involved in multiple biological processes via post-transcriptional regulation (Rajman and Schratt, 2017). Recent studies have discovered that Dicer-dependent miRs are involved in post-injury regulation of axon regeneration. After nerve damage in Dicer-knockout mice there was less regeneration of sensory nerves when assessed by various criteria (Wu et al., 2012). The miRs expression spectrum swings dramatically in the rodent SCI model. Some modulated miRs target RAGs which navigate axon regeneration (Li et al., 2016). Consequently, miRs are turning out to be crucial intrinsic epigenetic regulators that manipulate either central or peripheral axon regeneration post-injury. MiR-132 acts as a positive regulator, which promotes the axon extension during development, by suppressing its downstream target RasGTPase activator Rasa1 (Hachisuka et al., 2014). Interestingly, miR-132 is one of the up-regulated miRs post spinal cord ischemic injury (Yunta et al., 2012). A recent study in adult zebraf ish demonstrated that exogenous overexpression of miR-133b not only promotes the post-SCI axon regeneration but also largely improved the recovery of motor function by restricting the RhoA signaling pathway (Yu et al., 2011). In addition, miR-133b was proven to promote neurite outgrowth in both cortical neurons and PC12 line cells (Lu et al., 2015). In a rat ischemia-reperfusion SCI model, miR-210 was down-regulated more than 2-fold (Hu et al., 2013). Hu et al. (2016) found that inhibition of endogenous miR-210 in dorsal root ganglion impedes both in vitro and in vivo axon regeneration and that the inactivation of ephrin-A3 (EFNA3) successfully rescued axons from their inability to regenerate. Another study suggested that miR-210 overexpression promotes antiapoptosis and angiogenesis in a Wnt pathway-dependent manner, which improves the recovery of neurological function post-SCI (Ujigo et al., 2014). These results suggest that the SCI-induced miR-210 down-regulation is probably harmful to post-injury axon regeneration; modulation of miR-210 expression is expected to be a potential target for future SCI clinical trials. Additionally, the author’s group found that miR-26a expression promotes mammalian sensory axon regeneration by inhibiting glycogen synthase kinase 3β (GSK3β) mediated Smad1 inactivation (Jiang et al., 2015). Furthermore, miRs and certain epigenetic modifiers compose an epigenetic regulation feedback loop to operate gene expression jointly, such as the miR-138/SIRT1 negative feedback loop in the regulation of axon regeneration (Liu et al., 2013). As well as non-coding RNAs, certain RNA-binding proteins (RBP) also act as determinants during axon regeneration. Lin28 is an RBP that is highly conservative throughout evolution, which promotes the efficiency of human somatic cell reprogramming (Viswanathan and Daley, 2010). A recent study demonstrated that homologs of Lin28 and Lin28a/b are both required for axon regeneration in mature mammals. Significantly, Lin28a overexpression can largely promote axon regeneration in mature retinal ganglion cells of the CNS (Wang et al., 2018). Although the molecular mechanisms underlying axon regeneration and the pathogenesis of SCI have yet to be explored, the strong relationship between epigenetic factors and axon regeneration in the CNS has been adequately demonstrated in these studies. Consequently, a favorable orchestration of epigenetic factors and regulatory network is likely to turn sleeping mature neurons into a regenerative competent state, which will help develop promising novel therapeutic targets for SCI therapy.
Epigenetic Modifications and Secondary SCI
Epigenetic modifications and oligodendrocytes
Unlike transection in SCI animal models, the majority of clinical SCI patients are incompletely injured, which indicates that theoretically, the surviving axons in lesions are still capable of nerve conduction. However, the secondary SCI-induces widespread apoptosis of oligodendrocytes (OL) that leads to demyelination and axon degeneration, which eventually destroys the neural circuits. Therefore, promoting OL regeneration post-SCI is of great clinical significance for the recovery of neurological function. Even if stem cell transplantation could replace the cell loss post-SCI exogenously, limitations such as ethical principles and bio-safety still impede the clinical application of cell graft (Barnabé-Heider and Frisén, 2008). In rodent SCI models, although mature OLs could not regenerate, a gradual increase in the number of OLs was observed at the lesion site (Barnabé-Heider and Frisén, 2008). Based on this evidence, studies identified endogenous spinal cord stem cell and oligodendrocyte precursors (OPC) in the spinal cord as the main sources of OL accumulation post-injury. It is important to note that some epigenetic factors play critical roles during the recruitment and neurogenesis of endogenous spinal cord stem cells (Kameda et al., 2018; Koreman et al., 2018).
Enhancer of zeste homolog 2 (EZH2) is the catalytic subunit of histone methyltransferase polycomb repressive complex 2 (PRC2); it is capable of catalyzing the H3K27 trimethylation (Boyer et al., 2006). The lncOL1-mediated H3 trimethylation modulates OL lineage proliferation and differentiation (He et al., 2017). Another in vitro study suggested that EZH2 overexpression promotes OPC propagation and inhibits astrocyte expansion. In contrast, a reduced EZH2 intracellular expression decreased the percentage of OL and promoted AS generation (Sher et al., 2008). To some extent, H3K27me3 enrichment-induced OL differentiation seems to contradict its conservative epigenetic inhibitory roles. Although the specif ic mechanisms remain unknown, this paradoxical phenomenon might be related to the following reasons: (1) H3K27me3 and H3K4me3 form H3 bivalent domains in chromatin, and jointly regulate gene expression (Swigut and Wysocka, 2007). (2) H3K27me3 and H3K4me3 play domain repressive and permissive roles in gene regulation, respectively. In this case, certain inhibitory genes against OL differentiation might be blocked by EZH2-mediated H3K27me3. It will be interesting to discover the contributing genes in future studies. Postnatal OL cell-specif ic Dicer ablation leads to OL maturation associated phenotypes in the CNS, such as demyelination and neural degeneration. This finding indicates that miR-mediated gene expression regulation is critical for OL lineage differentiation (Shin et al., 2009; Dugas et al., 2010). MiR-219 was shown to inhibit OL proliferation-associated gene expressions, such as PDGFRα, Sox6, and FoxJ3. Dicer1 ablation significantly increased miR-219 expression in the early stages of OL differentiation. In rodent OPC, miR-219 overexpression rescues the Dicer1 ablation-induced OL differentiation disruption (Dugas et al., 2010). MiR-138 is expressed in the early phase of OL differentiation and down-regulated in mature OLs. The miR-138 overexpression in OPC prolongs the differentiation duration of OPC to OL (Emery and Lu, 2015). Total inhibition of HDAC activity blocks the early phase of OPC differentiation, rather than the later, onset of myelination phase (Conway et al., 2012). Recent studies have suggested that class I HDACs (HDAC1 and 2) are important for OPC differentiation in vitro (Shen et al., 2008). SIRT2, a class III HDAC dependent deacetylase, was discovered to be enriched in OLs and myelin sheath. A siRNA-induced SIRT2 knockdown not only reduces tubulin deacetylation, but promotes OPC differentiation (Li et al., 2007). Although the exact role of HDACs in OL lineage progression remains unexplored, the evidence suggests that it works in a stage-specif ic manner during OL-associated regulation.
Epigenetic modifications and astrocytes
Shortly after SCI, cellular events such as inf lammatory inf iltration, glial activation, and ependymal cell division are beneficial in the limitation of inf lammation and tissue damage repair. However, the secondary SCI-induced neuroinflammation cascades turn the spinal lesion into a hostile environment that impedes axon regeneration and the eventual survival of neurons. Reactive astrocytes are necessary factors for the post-SCI microenvironment but play roles of mixed benef it in SCI recovery (Cregg et al., 2014; Anderson et al., 2016). Emerging studies have suggested that epigenetic modifications regulate astrocyte proliferation and glia-associated astrocytes response. Recent studies have shown that miR-21 is expressed at low levels in an intact spinal cord but is elevated dramatically post SCI. A miR-21 inhibition promotes the response of reactive hypertrophic astrocytes, which encourage axon growth through the glial scar. In contrast, miR-21 overexpression represses the permissive axon response (Bhalala et al., 2012; Zhang et al., 2019). In addition, histone modifications are critical for the generation and migration of astrocytes. EZH2 expression in the subventricular zone of postnatal mice is required for neural stem cell (NSC) self-renewal and lineage differentiation toward the astrocyte lineage. EZH2 regulates the proliferation and lineage specif ication by mechanically repressing the expression of Ink4a or Oligo2, respectively (Hwang et al., 2014). The H3K9 demethylase, GASC1, is a marker of the distribution and migration of astrocytes, whereas abnormal astrocyte arrangements result in neurobehavioral phenotypes in Gasc1-hypomorphic mice (Sudo et al., 2016). Glial f ibrillary acidic protein (GFAP) is a type III intermediate f ilament in astrocytes. The GFAP expression prof iles correlates with injury responses and disease incidence in the CNS (Sticozzi et al., 2013; Clairembault et al., 2014). The inhibition of HDACs activity with Trichostatin-A represses the GFAP expression in astrocytes that results in incomplete astrocyte maturation. The intermediate filament network dysregulation and GFAP aggregation are possible molecular etiologies of some neurological diseases, such as Alexander’s disease (Schmidt et al., 2011).
Epigenetic modifications of microglia and immune system
Microglial polarization phenotypes are key determinants of the CNS inflammatory progression. In a naïve spinal cord, most microglials are M2 type (immunosuppressive type) which are beneficial for wound healing (Tang et al., 2014). However, secondary SCI turns microglials into the immune activated type (M1 type), which exaggerates the inflammatory response. Histone H3K27me3 demethylase Jumonji domain-containing protein 3 (JMJD3) was critical for microglial polarization. N9 microglial-specif ic inhibition of JMJD3 promotes M1 cell activation and inf lammation-induced neuron death. HDAC3 prevents M2 cell formation by inhibiting Th2 cytokines exposure. The deletion of HDAC3 significantly limits the inf lammatory disorders by enhancing M2 cell-induced alternative activation (Mullican et al., 2011). Additionally, a recent study demonstrated that HDAC3 inhibition promotes caspase-dependent apoptosis (Lankford et al., 2018). The miR-136-5p participates in the rodent post-SCI inf lammatory response by regulating inf lammatory cell inf iltration. Silencing of miR-136-5p reduces the expression of mediators such as interleukin-6, tumor necrosis factor alpha, and interferon-alpha, which ameliorates inf lammatory damage of the spinal cord (Yin et al., 2017; Deng et al., 2018; Lankford et al., 2018).
Apart from the neuroglia cell reaction, activation of the SCI-induced immune system also plays a critical role in pathological responses to post-injury (Vogelaar, 2016). Lymphocyte activation was detected in spinal cord tissue post-SCI, which could be negative or positive for SCI recovery (Ankeny et al., 2006). Continuous primary and secondary injury, not only in the spinal cord but in the blood-spinal cord barrier, result from severe damage. Consequently, self-antigens, such as myelin, phospholipids and cytokines, are exposed to either local lesions or peripheral blood. These exposures trigger an autoimmune reaction, exacerbating the tissue damage and eventual neuron loss (Schmidt et al., 2011). By transferring the MACS separated CD4+T cells into the SCI epicenter, the major histocompatibility class II (MHC II) knockout induced axon growth and post-SCI neural survival deficiency could be largely rescued (Walsh et al., 2015). The study concluded that, among these exogenous CD4+T lymphocytes, Th2 lymphocytes were the driving force for neuroprotection and functional recovery post-SCI.
Epigenetic modifications and blood-spinal cord barrier
The integrity of the blood-spinal cord barrier is crucial for the maintenance of the extracellular neuronal environment of the spinal cord. However, SCI-induced matrix metalloprotease activation results in blood-spinal cord barrier disruption that leads to catastrophic events such as inf lammatory inf iltration and neuronal apoptosis. Along with nuclear factor kappa-B, JMJD3 regulates the gene expressions of mmp-3/mmp-9 post-SCI. JMJD3 deletion not only decreases blood-spinal cord barrier permeability by matrix metalloprotease repression, but promotes the recovery of neurological function (Lee et al., 2016). Even though few direct studies support the theory that epigenetic modifications-based mechanisms regulate secondary SCI, the evidence suggests that the modulation of those epigenetic factors presents potential targets for the treatment of secondary SCI.
Epigenetic Biomarkers for SCI Prognosis
In view of the heterogeneous and complex nature of SCI, the outcomes and prognosis are generally hard to predict (Kwon et al., 2019). Consequently, a reliable system to forecast the prognosis and evaluate the effect of a treatment is important for the clinical treatment of SCI. Several prognostic SCI biomarkers, either in cerebrospinal fluid or peripheral blood, have been identified. Remarkably, some of the biomarker candidates are epigenetic factors which have been proved to be strongly associated with axon regeneration and glial activation post nerve injury (Yokobori et al., 2015; Rodrigues et al., 2018; Paim et al., 2019). The miRs may be accurate biomarkers because of their tissue specif icity and expression constancy. For some neurosurgery diseases, such as ossification of the posterior longitudinal ligament and low-grade gliomas, circulating miRs have already been identified as potential prognostic indicators (Xu et al., 2019). These injury-induced peripheral blood miRs modulations were first identified in a rodent SCI model (Nakanishi et al., 2010). They reported that miR-223 expression was up-regulated in the first few post-injury days. Conversely, there was a greater suppression of miR-124a expression in the first week after SCI. The bioinformatics significances of the miR modulation include responses such as inf lammatory activation and cell death (Nakanishi et al., 2010). Recent studies have recognized that miR-9 is a promising biomarker to evaluate the severity of acute-phase SCI in peripheral blood (Hachisuka et al., 2014; Paim et al., 2019). No blood-derived epigenetic biomarkers have been developed from human SCI patients, even though advantages such as minimal invasion and accuracy of miRs are very good. Certain specif ic miRs could potentially be used as prospective indicators for treatment and prognosis of SCI.
Exogenous Stimulation-Induced Epigenetic Modification
As well as acting as the driving force of SCI response, epigenetic modification states are also consequences of neuronal stimulations. The neuropathic pain initiated epigenetic alteration was demonstrated in a rodent peripheral nerve ligation model. The study found that sciatic nerve ligation triggers pro-inflammatory cytokine monocyte chemotactic protein-3 (MCP-3) up-regulation in the spinal cord. Analysis of spinal cord histone profiles post-injury detected diluted H3K27me3 at the promoter region of Mcp-3 (Descalzi et al., 2015). Recent studies demonstrated that exogenous transient neuronal activation triggered chromatin de-concentration, which largely enhances chromatin accessibility. In addition, the induced de-concentration was enriched at active enhancer regions, which were co-marked by permissive histone marks such as H3K4me1 and H3K27ac (Su et al., 2017). These results suggest that the neuronal stimulation-induced epigenetic modifications spread over the whole nerve system. This phenomenon might help explain the mechanism of rehabilitation therapies such as medium frequency electric stimulation from the epigenetic point of view. Accordingly, techniques such as electrophysiological stimuli and artif icial neuromodulator, which are competent to maneuver neuronal activation, will probably provide opportunities to regulate neurological function of the CNS via epigenetic modification mechanisms.
Conclusion and Perspective
SCI-induced neurological disability is predominantly attributed to failure of axons to regenerate in a spinal cord lesion. Recent studies have demonstrated that epigenetic regulations play pivotal roles in both axonal intrinsic regenerative responses and the regulation of extrinsic elements. Recent evidence has shown that epigenetic modifications and associated regulations are involved in key aspects of SCI recovery, such as axon regeneration, glial activation, inf lammatory response and endogenous NSC reprogram. These breakthroughs provide favorable candidates for SCI research and are promising targets for clinical SCI therapy (Goldman, 2016). These new discoveries can establish reliable epigenetic biomarker systems for forecasting SCI prognosis and clinical evaluation. With the improvement and optimization of bioinformatics databases, the implications for SCI-induced epigenetic changes could be further interpreted for clinical use.
Due to their small size and conserved nucleotide sequence, miRs act as pioneers in epigenetic related translational medicine research. With the development in chemical syntheses and drug delivery technologies, several clinical trials based on liposomes and antisense nucleotides have been launched. For instance, SPC3649, an anti-HCV drug by Santaris, based on miR-122 antisense nucleotide and PF-655 by Quark and Pfizer, which activates RTP801 gene expression, has been developed. The safety, tolerance, and therapeutic effect of these drugs have been fully confirmed in previous clinical trials. Even so, some outstanding challenges remain, such as the hybridization associated with off-target effects and delivery related concerns (Burnett and Rossi, 2012; Zhang et al., 2013; Li and Rana, 2014). Despite of the aforementioned progress, epigenetic mechanisms underlying SCI remain unclear. Significant efforts will be necessary for the exploration and verification of SCI associated epigenetic targets. With the deepening understanding of SCI molecular mechanisms, as well as the improvement in technology, epigenetics-based intervention approaches will def initely have broad and beneficial prospects in clinical SCI treatment.
Acknowledgments:We thank Ying Wang and Taylor Zhang from Changchun Ploy Developments Co., Ltd., China for language editing service.
Author contributions:Conceptualization: BYZ and Saijilafu; manuscript writing: BYZ and PYC; f igures drawing: BYZ and QSZ; manuscript revision: Saijilafu and YHZ. All authors reviewed and approved the final version of this manuscript.
Conf licts of interest:The authors declare no conf licts of interest.
Financial support:This work was supported by the National Natural Science Foundation of China, Nos. 81701225 (to BYZ), 81874254 (to PYC), 81571189 and 81772353 (to Saijilafu); the Excellent Youth Grant of Science and Technology Department of Jilin Province of China, No. 20190103077JH (to BYZ); the Bethune Project of Jilin University of China, No. 2015312 (to BYZ). The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-Non-Commercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewer:Wenrui Qu, Indiana University School of Medicine, USA.
Additional file:Open peer review report 1.
- 中国神经再生研究(英文版)的其它文章
- Recovery of an injured ascending reticular activating system with recovery from a minimally conscious state to normal consciousness in a stroke patient: a diffusion tensor tractography study
- The role of vascularization in nerve regeneration of nerve graft
- New insights into Wnt signaling alterations in amyotrophic lateral sclerosis: a potential therapeutic target?
- Advanced diffusion magnetic resonance imaging in patients with Alzheimer’s and Parkinson’s diseases
- Modulation of autophagy for neuroprotection and functional recovery in traumatic spinal cord injury
- Insights into platinum-induced peripheral neuropathy-current perspective

