Investigation of potential Shiga toxin producing Escherichia coli(STEC)associated with a local foodborne outbreak using multidisciplinary approaches
Kristen A.Lozink,Niket Jni,Jynthi Gngiredl,Ish Ptel,Christopher A.Elkins,Zonglin Hu,Prine A.Kssim,Roert A.Myers,Pongpn Lksnlmi,*
a Laboratories Administration,Maryland Department of Health and Mental Hygiene,1770 Ashland Ave.,Baltimore,MD 21205,United States
b Office of Applied Research and Safety Assessment,Center for Food Safety and Applied Nutrition,US Food and Drug Administration,8301 Muirkirk Rd.,Laurel,MD 20708,United States
c Winchester Engineering and Analytical Center,Office of Regulatory Affairs,US Food and Drug Administration,109 Holton St.,Winchester,MA 01890,United States
Abstract Shiga toxin producing Escherichia coli(STEC)outbreak is a public health concern as it can potentially cause a variety of clinical manifestations including diarrhea,hemorrhagic colitis and hemolytic uremic syndrome(HUS).However E.coli are generally innocuous commensal organisms,and there is a need to discriminate pathogenic from non-pathogenic isolates rapidly and accurately.In this study,we have used standard culture based methods and advanced molecular approaches to characterize E.coli in food in a local outbreak investigation.We show that the application of DNA based detection methods including real-time PCR and DNA microarray along with a traditional culture method can identify the organism implicated in an outbreak at the strain level for pathogenic potential.
Keywords:E.coli non-O157;Shiga toxin producing E.coli(STEC);RT-PCR;DNA microarray
1.Introduction
Many serotypes of Shiga toxin producingEscherichia coli(STEC) have emerged as a major cause of food-borne infections in the past 30 years[1–3].E.coliserotype O157:H7 STEC is considered one of the most important pathogens for public health concerns,classified as an adulterant by the US Food and Drug Administration and the USDA Food Safety and Inspection Service(FSIS)[4,5].Recently,it has also been recognized that a large number ofE.colinon O157:H7 serotypes can be responsible for manyE.colioutbreaks[6].A study at the Centers for Disease Control and Prevention(CDC)showed that from 1983 to 2002 approximately 70%of non-O157 STEC infections were caused by strains from one of the six major serogroups known as“Big Six”,including O26,O111,O45,O121,O103 and O145[7].STEC can potentially cause a variety of clinical manifestations including diarrhea,hemorrhagic colitis and hemolytic uremic syndrome(HUS)[8].Pathotypically,STEC can usually be classified as enterohemorrhagicE.coli(EHEC)[9].As such,the FDA Bacteriological Analytical Manual (BAM) protocol now requires analysts who perform food testing to screen samples for bothE.coliO157:H7 and non O157:H7 STEC[8,10].
For public health concerns,there is a need for rapid methods to identify,characterize and serotype pathogens associated with contaminated food during the surveillance and outbreak investigation.In addition to the traditional culture-based methods,several molecular assays including WGS,real-time (RT)PCR,DNA optical mapping.ELISA or whole genome microarray have emerged over the past several years [11,12].Our study focuses on using RT-PCR analyses and DNA microarray methods to assist an outbreak investigation of a localE.colinon-O157:H7 outbreak causing diarrhea in a few people from a food borne outbreak.
DNA microarray is one of the highly discriminatory sequence-based molecular approaches that can quickly and accurately identify the relatedness of organisms by presence/absence of genes in the pathogens.Low density microarray,targeting several major virulence genes has previously been proposed as a molecular tool to assess STEC [13].Since then,microarray technology has been improved with the development of bioinformatics as well as whole genome sequencing(WGS) technology [11].DNA microarray has been shown to provide advantage tools in the characterization of several major foodborne pathogens includingCronobactersp.[14],E.coli[11,15–17],Listeria monocytogenes[18–20]orClostridium botulinum[21].We have shown here that after the isolates were obtained,we were able to obtain the genomic information of the isolates within 48 h.The fast turnaround time is critically important not only for outbreak investigation,but is important for the regulatory perspective as well.
2.Materials and methods
2.1.Sample enrichment and biochemical tests
Our study was carried out in response to a possible localE.colinon-O157:H7 STEC outbreak investigation.We used two different approaches to characterize theE.colistrains implicated in this outbreak,including culture and molecular based methods.FDA-BAM was strictly followed as a standard method for culturingE.coliO157:H7 andE.colinon-O157:H7 STEC (BAM chapter 4a) [10].Several food samples implicated in this outbreak are shown in Table1.In brief,after the samples were received at our laboratory,microbiological examination was performed within 48 h of the collection,by enrichingtheminmodifiedbufferedpeptonewaterwithpyruvate(mBPWp) for 5 h at 37◦C,then supplemented with acriflavin,cefsulodin,and vancomycin (ACV) to the final concentration in mBPWp of 10 mg/L,10 mg/L and 8 mg/L,respectively.The sample enrichments were then incubated at 42◦C for a total of 18–24 h [10].The overnight sample enrichments were serially diluted in Butterfield’s phosphate buffer,with 0.5 mL of the 10−2through 10−4dilutions spread-plated in duplicate on Levineeosine methylene blue(EMB)agar and R&F®non-0157 STEC Chromogenic Plating Medium,a chromogenic agar that utilizes the chromogen,X-β-D-glucuronic acid to detect the enzyme,β-glucuronidase which is produced by 95%–98% ofE.coli,as well as someSalmonellaandShigellastrains.Addition of phenol red indicator,cellobiose and myo-inositol,differentiates these pathogens fromE.coli,and after 24 h incubation at 41–42◦C,E.colinon-O157 will appear as dark blue colonies with or without a clear ring.Further biochemical testing(MUG,indole)and confirmatory testing such as API20E(BioMérieux,Lyon,France) and ProlexTME.colinon-O157 latex kit (Pro-Lab Diagnostics,Round Rox,TX)were utilized to confirm that both isolates wereE.colinon-O157 serotype and of the O45 serogroup.
Table1 Samples that were collected for outbreak investigation.
2.2.RT-PCR methods
All sample enrichments were screened forE.colinon-O157:H7 STEC using both commercially available BAX-PCR(Dupont,Wilmington,USA) and an in-house real-time PCR[5,22,23].BAX PCR was performed according to the manufacturer’s recommendation.Real-time PCR for non-O157 STEC was performed using ABI7500 FAST Dx with the primers and probes specific to the Big Six group of theE.colinon-O157:H7 STEC shown in Tables 2A and 2B.
Primers and probes for RT-PCR(Tables 2A and 2B)obtained from Integrated DNA technologies (Coralville,IA),Biosearch Technologies(Novato,CA)and Invitrogen(Carlsbad,CA)were used to analyze the samples.Big six PCR panel real-time PCR was performed in a final volume of 25 μL in a 96-well plateusing the ABI7500 Fast DX real-time PCR system (Applied Biosystems,Foster City,CA).For each sample,seven singleplex PCR reactions were performed with six panel Big-6 analytes and a seventh 16S rRNA analyte based on the protocol received from United States Department of Agriculture,Food Safety and Inspection Service,Office of Public Health Science[24,25]with a few modifications.The final PCR reaction consisted of 1X TaqMan®Universal PCR mastermixII (Applied Biosystems,Foster City,CA) and the following:for O26,0.248 μmol/L of each of forward and reverse primers and 0.15 μmol/L probe;for O111,0.248 μmol/L of each of forward and reverse primers and 0.2 μmol/L probe; for O45,0.248 μmol/L of each of forward and reverse primers and 0.188 μmol/L probe; for O121,O103 and O145,0.248 μmol/L of each of forward and reverse primers and 0.2 μmol/L probe and for 16S,0.8 μmol/L of each of forward and reverse primers and 0.2 μmol/L probe respectively.Five microliters of extracted DNA template was added to each reaction in a total volume of 25 μL.Cycling conditions were as follows,95◦C for 10 min,followed by 45 cycles of denaturation at 95◦C for 15 s and annealing at 59◦C for 60 s.A sample was reported as positive in a valid run if any of the serogroup analyte was detected by PCR(Ct≤38.0)and negative if all of the serogroup analytes were not detected by PCR.

Table2A Primers sequences for PCR used to screen E.coli non-O157:H7 STEC.

Table2B TaqMan probes used to screen E.coli non-O157:H7 STEC.

Table3 Ct values from all of the tested primers.
2.3.Microarray analysis
High-density Affymetrix custom microarray designed forE.coli[26]was used to further characterize the two isolates.The isolates were grown in Gram negative broth (Fisher Scientific,Pittsburgh,PA)at 35◦C overnight and total DNA from 0.5 mL of culture was extracted using a DNeasy kit (Qiagen,Venlo,Netherlands).Genomic DNA was digested by RQ1 RNA free DNaseI for 1 min at 37◦C to obtain an average fragment size of ~200 bp.The DNA fragments were then labeled with biotin using 30 U of recombinant terminal deoxynuclotidyl transferase(rTdT),(Promega Corp,WI) for 4 h.The labeled fragments were hybridized onto the FDA-ECID gene chip,washed,stained and scanned using Affymetrix GeneAtlas instrument following Lacher et al.[26]and manufacturer’s recommendation.Mas5.0 and Robust Multi Array (RMA) were performed as described previously[19,26–29].
3.Results and discussion
We first screened all of the enrichments using BAX PCR forE.coliO157 and non O157 STEC.The results indicated that none of the samples hasE.coliO157 strains.However,the raw chicken sample(002)was positive from the originalE.colinon O157 STEC screening panel.Using the two recommended BAX PCR panels,we subsequently tested the sample to identify the Big Six group,finding that the sample was positive forE.coliserogroup O45.Following the FDA-BAM protocol,we were able to isolate twoE.colinon-O157:H7 isolates from a raw chicken sample(002)(Table1).Samples 002,from raw chicken which was positive forE.colinon O157 STEC screening,was further screened on Big-6 PCR panel.We found that the 002 broth as well as the isolates from which the two 002 isolates were obtained were positive for O45 serogroup (Fig.1A).Interestingly,in addition to the positivity to the O45 group,sample 002 broth was also positive for O111 serogroup(Fig.1B).This suggests that sample 002 may contaminated with both serogroups,O45 and O111.However,theCtvalues for O111 is much higher than that of O45(Table3)suggesting that O45 may be predominant in the population and the culture based method may not be sensitive enough to enable us to isolate both strains.This indicates the advantage of molecular detection.Further biochemical testing (MUG,indole) and confirmatory testing with API20E and ProlexTME.colinon-O157 latex kit strongly confirmed that both isolates wereE.colinon-O157 serotype and of the O45 serogroup.
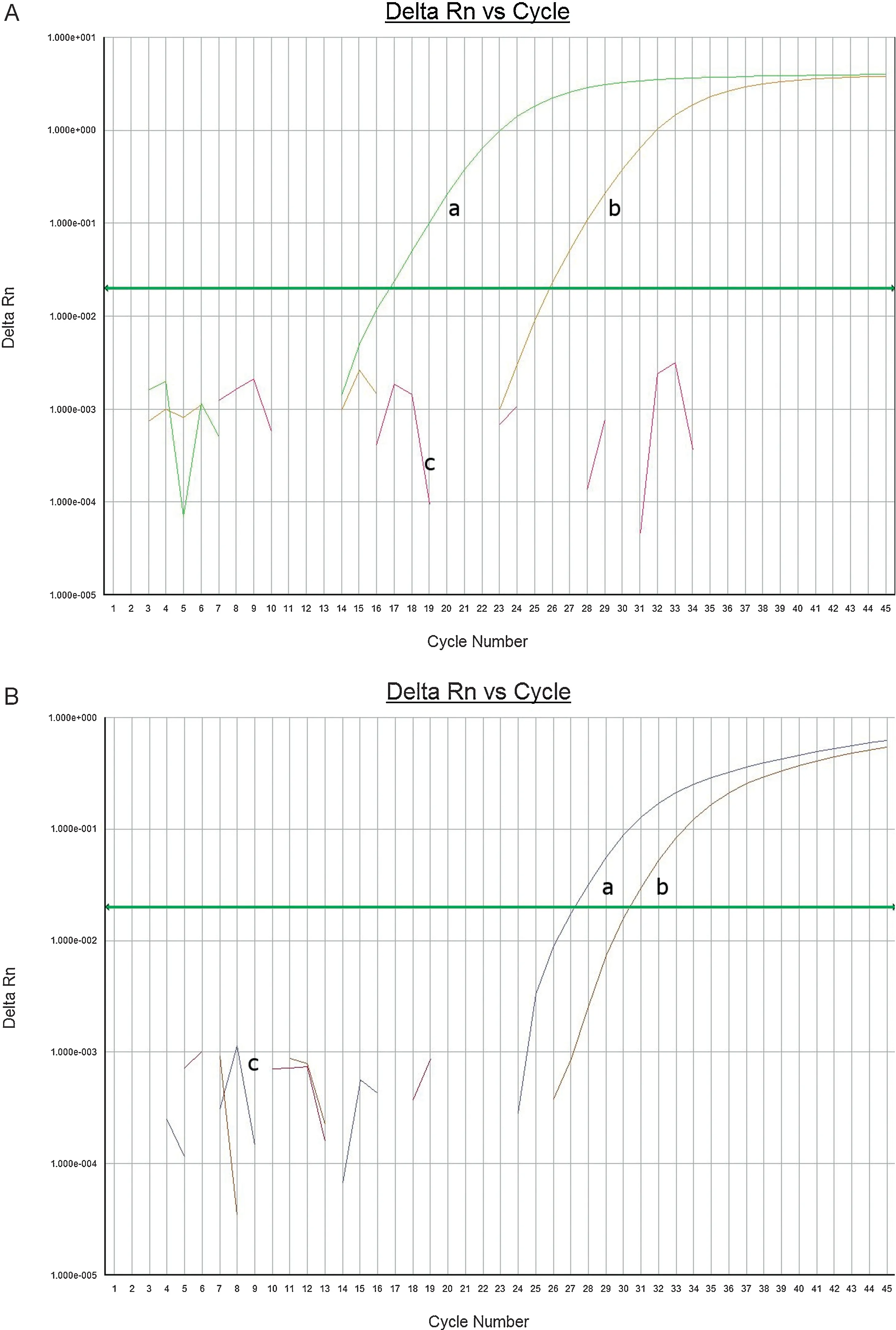
Fig.1.(A)O4A5 Analyte PCR amplification curve of 002 Broth(a),O45 standard DNA(b)wells.NTC(c)suggests no amplification as expected.(B)O111 analyte PCR amplification curve of 002 Broth(a),O111 standard DNA(b)wells.NTC(c)shown in brown suggests no amplification as expected.

Fig.2.Comparison of probe-set intensity between isolates A and B.
Genomic DNA extracted from these two isolates were subjected to Microarray analysis.The analyzed results were obtained within approximately 48 h after isolates had been received.In addition to the fast turnaround and less handson time relative to WGS,using microarray technology in the real-time outbreak investigation can be very important as it provides perspective on strains and the pathotype relationships[11].DNA microarray analysis can be relatively quick to perform using R and Bioconductor software,available freely at https://www.bioconductor.org/.In general,there are two main methods,RMA and MAS5.0,for microarray analysis.In brief,RMA result reveals the intensity oflabeled target DNA from the entire genome as they bind to the probes.MAS5.0 algorithm,on the other hand,statistically calculates the intensity of mismatched and matched probe/targets and reports the results as gene presence/absence[19].
We used both methods to characterize these isolates.The relatedness of these two isolates was determined by RMA and the results clearly indicated that the two isolates are almost identical(Fig.2).
Mas5.0 was used to obtain the gene content and serotype of the isolates.Traditionally,serotyping ofE.coliwas performed using pools of polyclonal antisera against these O-and H-antigens.However,the process is labor-intensive and time consuming.An alternative method is a molecular serotyping by detecting the presence of specific antigenic gene sequences.In this case,O-serotype is identified by the presence of a specificwzxandwzygenes whereas H-serotype is fromfliCgenes[26,30,31].As the MAS5.0 analysis can be used to identify gene contents,we further examined the genes that encode for O-(wzx,wzyandwzm) and H- (fliC) antigens.The microarray results indicated that these isolates harborwzx45wzy45 andfliC8 with all otherwzx,wzy,wzmandfliCgenes absent.This result confirmed that the isolates areE.coliO45:H8.All of thestxandeaegenes are absent suggesting that these two isolates are not pathogenic.In addition,some of other virulence genes encoding virulence proteins such as bundle-forming pilus(bfp)found in EnteropathogenicE.coli,or hemolysinA(hlyA)were also absent in both O45 isolates.
Environmental samples may contain mixed populations of STEC strains,many of which could share O-type antigens with a variety of H-antigens.It has been suggested that identifyingfliC gene type may be helpful for detection of pathogenic STEC strains.Several H-types including H-,H2,H7,H8,H11,H12,H16,H19,H21,H25 and H28 are commonly associated with STEC strains.However,O serogroups when combined with different H antigens may contribute to different pathogenicity of the strains.For example,Serogroup O104 with the H7 or H21 can be more pathogenic than other combinations[32].As a result,using commercial kits for identification of O-group to confirm the presence of pathogenic STEC may result in false positive [4].Our results showed that there were multiple serotypes in the samples;the commercial and custom PCR screening results indicated that the samples were contaminated with two O-serotypes belonging to the Big Six group of STEC,O45 and O111.Serotype O45 combined with H2 appears to be a pathogenic strain as it harbors Shiga toxin 2 called Stx2f[33].Our isolates,on the other hand,were identified as O45:H8,an O:H serotype combination that may not appear to be pathogenic.The microarray analysis also revealed that both isolates did not possess any of thestxandeaegenes.This result also suggested that O:H serotype may reflect pathogenicity ofE.colistrains.The positive result from BAX PCR forstxgenes used to screenE.colinon O157:H7 STEC possibly came from the presence of O111 strains.However,due to the low population of O111 in the sample relative to O45 as shown in RT-PCR,the BAM culture method may not have enough sensitivity to allow the strain that may be responsible for the outbreak to grow.Our study strongly suggested while culture methods remain as gold standards for organism identification in foods,modern methods such as real time PCR and microarrays if used would provide concordant and additional information which could easily be escaped by culture methods due to lower limits of detection.In conclusion multidisciplinary approaches should be adopted during the outbreak investigation to help identify and characterize pathogens accurately.
Acknowledgements
We thank Kyle Shannon,Office of Food Protection,MDDHMH,for collecting,coordinating and providing all of the food samples from this outbreak.We also thank Keith A.Lampel for comments and critically reading the manuscript.
The views presented in this article do not necessarily reflect those of the Food and Drug Administration and MD Department of Health and Mental Hygiene.
- 食品科学与人类健康(英文)的其它文章
- Assessment of food toxicology
- Genetically modified foods:A critical review of their promise and problems
- Genetically modified foods in China and the United States:A primer of regulation and intellectual property protection
- Risk factors in street food practices in developing countries:A review
- Review of Finger millet(Eleusine coracana(L.)Gaertn):A power house of health benefiting nutrients
- Prevalence of fumonisin producing Fusarium verticillioides associated with cereals grown in Karnataka(India)

